Abstract
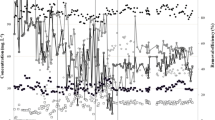
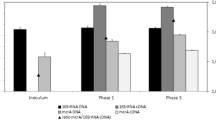
This study investigated methanogenic communities involved in degradation of tetramethylammonium hydroxide (TMAH) in three full-scale bioreactors treating TMAH-containing wastewater. Based on the results of terminal-restriction fragment-length polymorphism (T-RFLP) and quantitative PCR analyses targeting the methyl-coenzyme M reductase alpha subunit (mcrA) genes retrieved from three bioreactors, Methanomethylovorans and Methanosarcina were the dominant methanogens involved in the methanogenic degradation of TMAH in the bioreactors. Furthermore, batch experiments were conducted to evaluate mcrA messenger RNA (mRNA) expression during methanogenic TMAH degradation, and the results indicated that a higher level of TMAH favored mcrA mRNA expression by Methansarcina, while Methanomethylovorans could only express considerable amount of mcrA mRNA at a lower level of TMAH. These results suggest that Methansarcina is responsible for methanogenic TMAH degradation at higher TMAH concentrations, while Methanomethylovorans may be important at a lower TMAH condition.





Similar content being viewed by others
References
Angelidaki I, Ahring BK (1993) Thermophilic anaerobic digestion of livestock waste: the effect of ammonia. Appl Microbiol Biotechnol 38:560–564
APHA (1995) Standard methods for the examination of water and wastewater, 19th edn. APHA American Public Health Association, Washington, DC
Asakawa S, Sauer K, Liesack W, Thauer RK (1998) Tetramethylammonium: coenzyme M methyltransferase system from Methanococcoides sp. Arch Microbiol 170:220–226
Chang KF, Yang SY, You HS, Pan JR (2008) Anaerobic treatment of tetramethylammonium hydroxide (TMAH) containing wastewater. IEEE Trans Semicond Manuf 21:486–491
Chaudhary PP, Brablcová L, Buriánková I, Rulík M (2013) Molecular diversity and tools for deciphering the methanogen community structure and diversity in freshwater sediments. Appl Microbiol Biotechnol 97:7553–7562
Collins RE, Rocap G (2007) REPK: an analytical Web server to select restriction endonucleases for terminal restriction fragment length polymorphism analysis. Nucleic Acids Res 35:W58–W62
Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Ba¨umer S, Jacobi C, Brüggemann H, Lienard T, Christmann A, Bo¨meke M, Steckel S, Bhattacharyya A, Lykidis A, Overbeek R, Klenk HP, Gunsalus RP, Fritz HJ, Gottschalk G (2002) The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol 4:453–461
Ferry JG (1993) Methanogenesis, ecology, physiology, biochemistry and genetics. Springer, New York
Ferry JG (1999) Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol Rev 23:13–38
Freitag TE, Prosser JI (2009) Correlation of methane production and functional gene transcriptional activity in a peat soil. Appl Environ Microbiol 75:6679–6687
Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, Mc-Ewan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Graham DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, Conway de Macario E, Ferry FG, Jarrell KF, Jing H, Macario AJ, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B (2002) The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Res 12:532–542
Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR (1996) Isolation and identification of methanogen-specific DNA from blanket bog feat by PCR amplification and sequence analysis. Appl Environ Microbiol 62:668–675
Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF (2003) Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing Archaea. Appl Environ Microbiol 69(9):5483–5491
Hippe H, Caspari D, Fiebig K, Gottschalk G (1979) Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci 76:494–498
Hirano K, Okamura J, Taira T, Sano K, Toyoda A, Ikeda M (2001) An efficient treatment technique for TMAH wastewater by catalytic oxidation. IEEE Trans Semicond Manuf 14:202–206
Hu TH, Whang LM, Liu PWG, Hung YC, Chen HW, Lin LB, Chen CF, Chen SK, Hsu SF, Shen W, Fu R, Hsu R (2012) Biological treatment of TMAH (tetra-methyl ammonium hydroxide) in a full-scale TFT-LCD wastewater treatment plant. Bioresour Technol 113:303–310
Jiang B, Parshina S, van Doesburg W, Lomans B, Stams A (2005) Methanomethylovorans thermophila sp. nov., a thermophilic, methylotrophic methanogen from an anaerobic reactor fed with methanol. Int J Syst Evol Microbiol 55:2465–2470
Karakashev D, Batstone DJ, Angelidaki I (2005) Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl Environ Microbiol 71:331–338
Karakashev D, Batstone DJ, Trably E, Angelidaki I (2006) Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl Environ Microbiol 72:5138–5141
Konig H, Stetter K (1982) Isolation and characterization of Methanolobus tindarius sp. nov., a coccoid methanogen growing only on methanol and methylamines. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1:478–490
Lei CN, Whang LM, Chen PC (2010) Biological treatment of thin-film transistor liquid crystal display (TFT-LCD) wastewater using aerobic and anoxic/oxic sequencing batch reactors. Chemosphere 81:57–64
Liu Y, Boone DR, Sleat R, Mah RA (1985) Methanosarcina mazei LYC, a new methanogenic isolate which produces a disaggregating enzyme. Appl Environ Microbiol 49:608–613
Lomans BP, Maas R, Luderer R, Op den Camp HJM, Pol A, van der Drift C, Vogels GD (1999) Isolation and characterization of Methanomethylovorans hollandica gen. nov., sp. nov., isolated from freshwater sediment, a methylotrophic methanogen able to grow on dimethyl sulfide and methanethiol. Appl Environ Microbiol 65:3641–3650
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371
Lueders T, Friedrich MW (2003) Evaluation of PCR amplification bias by terminal restriction fragment length polymorphism analysis of small-subunit rRNA and mcrA genes by using defined template mixtures of methanogenic pure cultures and soil DNA extracts. Appl Environ Microbiol 69:320–326
Lueders T, Chin KJ, Conrad R, Friedrich M (2001) Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ Microbiol 3:194–204
Luton PE, Wayne JM, Sharp RJ, Riley PW (2002) The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521–3530
Lyimo TJ, Pol A, Jetten MSM, den Camp HJMO (2009) Diversity of methanogenic archaea in a mangrove sediment and isolation of a new Methanococcoides strain. FEMS Microbiol Lett 291:247–253
Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR (2006) The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J Bacteriol 188:7922–7931
Mrklas O, Chu A, Lunn S (2003) Determination of ethanolamine, ethylene glycol and triethylene glycol by ion chromatography for laboratory and field biodegradation studies. J Environ Monitor 5:336–340
Patel G, Sprott G (1990) Methanosaeta concilii gen nov., sp. nov.,(Methanothrix concilii) and Methanosaeta thermoacetophila nom. rev., comb. nov. Int J Syst Bacteriol 40:79–82
Peck JE (2010) Multivariate analysis for community ecologists: step-by-step using PC-ORD. MjM Software Design, Oregon
Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J, Glöckner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196
Simankova MV, Kotsyurbenko OR, Lueders T, Nozhevnikova AN, Wagner B, Conrad R, Friedrich MW (2003) Isolation and characterization of new strains of methanogens from cold terrestrial habitats. Syst Appl Microbiol 26:312–318
Singh N, Kendall MM, Liu Y, Boone DR (2005) Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica. Int J Syst Bacteriol 55:2531–2538
Sowers KR, Ferry JG (1983) Isolation and characterization of a methylotrophic marine methanogen, Methanococcoides methylutens gen. nov., sp. nov. Appl Environ Microbiol 45:684–690
Sowers KR, Baron SF, Ferry JG (1984) Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl Environ Microbiol 47:971–978
Springer E, Sachs MS, Woese CR, Boone DR (1995) Partial gene sequences for the A subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int J Syst Bacteriol 45:554–559
Steinberg LM, Regan JM (2008) Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl Environ Microbiol 74:6663–6671
Steinberg LM, Regan JM (2009) mcrA-Targeted real-time quantitative PCR method to examine methanogen communities. Appl Environ Microbiol 75(13):4435–4442
Tanaka K (1994) Anaerobic degradation of tetramethylammonium by a newly isolated marine methanogen. J Ferment Bioeng 78:386–388
Urakami T, Araki H, Kobayashi H (1990) Isolation and identification of tetramethylammonium-biodegrading bacteria. J Ferment Bioeng 70:41–44
Watanabe T, Kimura M, Asakawa S (2009) Distinct members of a stable methanogenic archaeal community transcribe mcrA genes under flooded and drained conditions in Japanese paddy field soil. Soil Biol Biochem 41:276–285
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci 87:4576–4579
Wu CL, Su SB, Chen JL, Lin HJ, Guo HR (2008) Mortality from dermal exposure to tetramethylammonium hydroxide. J Occup Health 50:99–102
Yuan YL, Conrad R, Lu YH (2011) Transcriptional response of methanogen mcrA genes to oxygen exposure of rice field soil. Environ Microbiol Rep 3:320–328
Acknowledgments
The authors would like to acknowledge the financial support from the Bureau of Energy, Ministry of Economic Affairs Energy Technology Program for Academia under grant no. 102-D0613, the Ministry of Education of Taiwan under grant for the Top University Project to the National Cheng Kung University, and partially financial support from the Innolux Corporation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Whang, LM., Hu, TH., Liu, PW.G. et al. Molecular analysis of methanogens involved in methanogenic degradation of tetramethylammonium hydroxide in full-scale bioreactors. Appl Microbiol Biotechnol 99, 1485–1497 (2015). https://doi.org/10.1007/s00253-014-6058-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-6058-z