Abstract
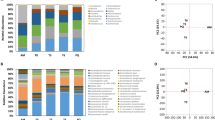
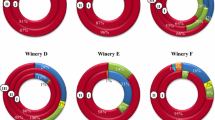
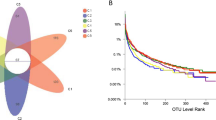
Grape marc used for the production of distilled beverages undergoes prolonged storage which allows alcoholic fermentation to occur. Harsh conditions including low pH, limited oxygen and nutrients, temperature fluctuations, and high ethanol concentrations imposed by that environment create a strong selective pressure on microorganisms. A detailed characterization of the bacterial community during two time points of the fermentation process was performed using high-throughput sequencing of the V3–V6 16S rDNA hypervariable regions. The results revealed a marked reduction in the number of bacterial species after 30 days of incubation and made it possible to identify those species that are able to grow in that extreme environment. The genome sequence of Lactobacillus fabifermentans, one of the dominant species identified, was then analyzed using shotgun sequencing and comparative genomics. The results revealed that it is one of the largest genomes among the Lactobacillus sequenced and is characterized by a large number of genes involved in carbohydrate utilization and in the regulation of gene expression. The genome was shaped through a large number of gene duplication events, while lateral gene transfer contributed to a lesser extent with respect to other Lactobacillus species. According to genomic analysis, its carbohydrate utilization pattern and ability to form biofilm are the main genetic traits linked to the adaptation the species underwent permitting it to grow in fermenting grape marc.








Similar content being viewed by others
References
Agogué H, Lamy D, Neal PR, Sogin ML, Herndl GJ (2011) Water mass‐specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol 20:258–274
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75
Bae S, Fleet G, Heard G (2006) Lactic acid bacteria associated with wine grapes from several Australian vineyards. J Appl Microbiol 100:712–727
Baker G, Smith J, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555
Bokulich NA, Joseph CL, Allen G, Benson AK, Mills DA (2012) Next-generation sequencing reveals significant bacterial diversity of botrytized wine. PLoS ONE 7:e36357
Borneman AR, McCarthy JM, Chambers PJ, Bartowsky EJ (2012) Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways. BMC Genomics 13:373
Bose M, Barber RD (2006) Prophage Finder: a prophage loci prediction tool for prokaryotic genome sequences. In Silico Biol 6:223–227
Botella C, Diaz A, De Ory I, Webb C, Blandino A (2007) Xylanase and pectinase production by Aspergillus awamori on grape pomace in solid state fermentation. Process Biochem 42:98–101
Bovo B, Fontana F, Giacomini A, Corich V (2011) Effects of yeast inoculation on volatile compound production by grape marcs. Ann Microbiol 61:117–124
Bovo B, Nardi T, Fontana F, Carlot M, Giacomini A, Corich V (2012) Acidification of grape marc for alcoholic beverage production: effects on indigenous microflora and aroma profile after distillation. Int J Food Microbiol 152:100–106
Bratlie M, Johansen J, Sherman B, Lempicki R, Drabløs F (2010) Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics 11:588
Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, Eddy SR, Gardner PP, Bateman A (2013) Rfam 11.0: 10 years of RNA families. Nucleic Acids Res 41:D226–D232
Cai H, Thompson R, Budinich MF, Broadbent JR, Steele JL (2009) Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol 1:239
Callanan M, Kaleta P, O'Callaghan J, O’Sullivan O, Jordan K, McAuliffe O, Sangrador-Vegas A, Slattery L, Fitzgerald GF, Beresford T (2008) Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J Bacteriol 190:727–735
Campanaro S, De Pascale F, Telatin A, Schiavon R, Bartlett DH, Valle G (2012) The transcriptional landscape of the deep-sea bacterium Photobacterium profundum in both a toxR mutant and its parental strain. BMC Genomics 13:567
Chassy BM (1985) Prospects for improving economically significant Lactobacillus strains by ‘genetic technology’. Trends Biotechnol 3:273–275
Cleenwerck I, De Vos P (2008) Polyphasic taxonomy of acetic acid bacteria: an overview of the currently applied methodology. Int J Food Microbiol 125:2–14
Cleenwerck I, Vandemeulebroecke K, Janssens D, Swings J (2002) Re-examination of the genus Acetobacter, with descriptions of Acetobacter cerevisiae sp. nov. and Acetobacter malorum sp. nov. Int J Syst Evol Microbiol 52:1551–1558
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen A, McGarrell D, Marsh T, Garrity GM (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Collins M, Rodrigues U, Ash C, Aguirre M, Farrow J, Martinez-Murcia A, Phillips B, Williams A, Wallbanks S (1991) Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett 77:5–12
Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinforma 11:485
Darling AE, Mau B, Perna NT (2010) progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147
Davis C, Wibowo D, Eschenbruch R, Lee T, Fleet G (1985) Practical implications of malolactic fermentation: a review. Am J Enol Vitic 36:290–301
De Bruyne K, Camu N, De Vuyst L, Vandamme P (2009) Lactobacillus fabifermentans sp. nov. and Lactobacillus cacaonum sp. nov., isolated from Ghanaian cocoa fermentations. Int J Syst Evol Microbiol 59:7–12
De Francisci D, Campanaro S, Kornfeld G, Siddiqui KS, Williams TJ, Ertan H, Treu L, Pilak O, Lauro FM, Harrop SJ (2011) The RNA polymerase subunits E/F from the Antarctic archaeon Methanococcoides burtonii bind to specific species of mRNA. Environ Microbiol 13:2039–2055
De Pina C, Hogg T (1999) Microbial and chemical changes during the spontaneous ensilage of grape pomace. J Appl Microbiol 86:777–784
De Rosa T, Castagner R (1994) Tecnologia delle grappe e dei distillati d’uva. Edagricole, Bologna, Italy
Diep DB, Straume D, Kjos M, Torres C, Nes IF (2009) An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides 30:1562–1574
Douillard FP, Ribbera A, Kant R, Pietilä TE, Järvinen HM, Messing M, Randazzo CL, Paulin L, Laine P, Ritari J (2013) Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG. PLoS Genet 9:e1003683
Dumbrepatil A, Adsul M, Chaudhari S, Khire J, Gokhale D (2008) Utilization of molasses sugar for lactic acid production by Lactobacillus delbrueckii subsp. delbrueckii mutant Uc-3 in batch fermentation. Appl Environ Microbiol 74:333–335
Freeman WM, Walker SJ, Vrana KE (1999) Quantitative RT-PCR pitfalls and potential. Biotechniques 26:112–125
Galeote V, Novo M, Salema-Oom M, Brion C, Valério E, Gonçalves P, Dequin S (2010) FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high-affinity fructose/H+ symporter. Microbiology 156:3754–3761
Gomez-Alvarez V, Teal TK, Schmidt TM (2009) Systematic artifacts in metagenomes from complex microbial communities. ISME J 3:1314–1317
Grissa I, Vergnaud G, Pourcel C (2007) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57
Guergoletto KB, Magnani M, Martin JS, Andrade, de Jesus Andrade CG, Garcia S (2010) Survival of Lactobacillus casei (LC-1) adhered to prebiotic vegetal fibers. Innov Food Sci Emerg Technol 11:415–421
Hofvendahl K, Hahn-Hägerdal B (1997) L-lactic acid production from whole wheat flour hydrolysate using strains of Lactobacilli and Lactococci. Enzyme Microb Technol 20:301–307
Huber B, Eberl L, Feucht W, Polster J (2003) Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z Naturforsch C 58:879–884
Hugenholtz P (2002) Exploring prokaryotic diversity in the genomic era. Genome Biol 3:reviews0003-reviews0003.8
Ilmén M, Koivuranta K, Ruohonen L, Suominen P, Penttilä M (2007) Efficient production of L-lactic acid from xylose by Pichia stipitis. Appl Environ Microbiol 73:117–123
Ingram LO, Dombek KM (1987) On the evolution of alcohol tolerance in microorganisms. Perspectives in Biotechnology. Springer, New York, NY, In, pp 131–138
Jordan IK, Makarova KS, Spouge JL, Wolf YI, Koonin EV (2001) Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res 11:555–565
Joyeux A, Lafon-Lafourcade S, Ribéreau-Gayon P (1984) Evolution of acetic acid bacteria during fermentation and storage of wine. Appl Environ Microbiol 48:153–156
Kalathenos P, Baranyi J, Sutherland JP, Roberts TA (1995) A response surface study on the role of some environmental factors affecting the growth of Saccharomyces cerevisiae. Int J Food Microbiol 25:63–74
Kato S, Ishihara T, Hemmi H, Kobayashi H, Yoshimura T (2011) Alterations in D-amino acid concentrations and microbial community structures during the fermentation of red and white wines. J Biosci Bioeng 111:104–108
Kawarai T, Furukawa S, Ogihara H, Yamasaki M (2007) Mixed-species biofilm formation by lactic acid bacteria and rice wine yeasts. Appl Environ Microbiol 73:4673–4676
Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW (2003) Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A 100:1990–1995
Korakli M, Pavlovic M, Gänzle MG, Vogel RF (2003) Exopolysaccharide and kestose production by Lactobacillus sanfranciscensis LTH2590. Appl Environ Microbiol 69:2073–2079
Kubota H, Senda S, Tokuda H, Uchiyama H, Nomura N (2009) Stress resistance of biofilm and planktonic Lactobacillus plantarum subsp. plantarum JCM 1149. Food Microbiol 26:592–597
Kummerfeld SK, Teichmann SA (2006) DBD: a transcription factor prediction database. Nucleic Acids Res 34:D74–D81
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Laslett D, Canbäck B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Lebeer S, Vanderleyden J, De Keersmaecker SC (2010) Adaptation factors of the probiotic Lactobacillus rhamnosus GG. Benef Microbes 1:335–342
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Lisdiyanti P, Kawasaki H, Seki T, Yamada Y, Uchimura T, Komagata K (2000) Systematic study of the genus Acetobacter with descriptions of Acetobacter indonesiensis sp. nov., Acetobacter tropicalis sp. nov., Acetobacter orleanensis (Henneberg 1906) comb. nov., Acetobacter lovaniensis (Frateur 1950) comb. nov., and Acetobacter estunensis (Carr 1958) comb. nov. J Gen Appl Microbiol 46:147–165
Lisdiyanti P, Navarro RR, Uchimura T, Komagata K (2006) Reclassification of Gluconacetobacter hansenii strains and proposals of Gluconacetobacter saccharivorans sp. nov. and Gluconacetobacter nataicola sp. nov. Int J Syst Evol Microbiol 56:2101–2111
Llaubères R, Richard B, Lonvaud A, Dubourdieu D, Fournet B (1990) Structure of an exocellular β-D-glucan from Pediococcus sp., a wine lactic bacteria. Carbohydr Res 203:103–107
Lorenzo F, Viviana C, Alessio G, Marina B, Sergio C (2013) Grape marcs as unexplored source of new yeasts for future biotechnological applications. World J Microbiol Biotechnol 29(9):1–12
Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115
Manitto P, Chialva F, Speranza G, Rinaldo C (1994) Absolute stereochemistry and enantiomeric excess of 2-butanol in distilled spirits of different origin. J Agric Food Chem 42:886–889
Maragkoudakis PA, Nardi T, Bovo B, D’Andrea M, Howell KS, Giacomini A, Corich V (2013) Biodiversity, dynamics and ecology of bacterial community during grape marc storage for the production of grappa. Int J Food Microbiol 162:143–151
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229
Medina de Figueroa R, Alvarez F, de Ruiz P, Holgado A, Oliver G, Sesma F (2000) Citrate utilization by homo-and heterofermentative lactobacilli. Microbiol Res 154:313–320
Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A (2008) The metagenomics RAST server-a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma 9:386
Molenaar D, Bringel F, Schuren FH, de Vos WM, Siezen RJ, Kleerebezem M (2005) Exploring Lactobacillus plantarum genome diversity by using microarrays. J Bacteriol 187:6119–6127
Nielsen DS, Teniola O, Ban-Koffi L, Owusu M, Andersson T, Holzapfel W (2007) The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol 114:168–186
Nykänen L (1986) Formation and occurrence of flavor compounds in wine and distilled alcoholic beverages. Am J Enol Vitic 37:84–96
O’Donnell MM, O’Toole PW, Ross RP (2013) Catabolic flexibility of mammalian-associated lactobacilli. Microb Cell Fact 12:48
Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304
Oh H, Wee Y, Yun J, Ho Han S, Jung S, Ryu H (2005) Lactic acid production from agricultural resources as cheap raw materials. Bioresour Technol 96:1492–1498
Ohkouchi Y, Inoue Y (2006) Direct production of L-lactic acid from starch and food wastes using Lactobacillus manihotivorans LMG18011. Bioresour Technol 97:1554–1562
Oliver JD (2005) The viable but nonculturable state in bacteria. J Microbiol 43:93–100
Papalexandratou Z, Vrancken G, De Bruyne K, Vandamme P, De Vuyst L (2011) Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol 28:1326–1338
Parks DH, Beiko RG (2010) Identifying biologically relevant differences between metagenomic communities. Bioinformatics 26:715–721
Patel MA, Ou MS, Harbrucker R, Aldrich HC, Buszko ML, Ingram LO, Shanmugam K (2006) Isolation and characterization of acid-tolerant, thermophilic bacteria for effective fermentation of biomass-derived sugars to lactic acid. Appl Environ Microbiol 72:3228–3235
Pittet V, Abegunde T, Marfleet T, Haakensen M, Morrow K, Jayaprakash T, Schroeder K, Trost B, Byrns S, Bergsveinson J (2012) Complete genome sequence of the beer spoilage organism Pediococcus claussenii ATCC BAA-344 T. J Bacteriol 194:1271–1272
Pittet V, Phister TG, Ziola B (2013) Transcriptome sequence and plasmid copy number analysis of the brewery isolate Pediococcus claussenii ATCC BAA-344 T during growth in beer. PLoS ONE 8:e73627
Pozo-Bayón M, G-Alegría E, Polo M, Tenorio C, Martín-Álvarez P, Calvo De La Banda MT, Ruiz-Larrea F, Moreno-Arribas M (2005) Wine volatile and amino acid composition after malolactic fermentation: effect of Oenococcus oeni and Lactobacillus plantarum starter cultures. J Agric Food Chem 53:8729–8735
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J (2012) The Pfam protein families database. Nucleic Acids Res 40:D290–D301
Pushker R, Mira A, Rodríguez-Valera F (2004) Comparative genomics of gene-family size in closely related bacteria. Genome Biol 5:R27
Quere F, Deschamps A, Urdaci M (1997) DNA probe and PCR‐specific reaction for Lactobacillus plantarum. J Appl Microbiol 82:783–790
Ribéreau-Gayon P, Dubourdieu D, Donèche B, Lonvaud A (2006) Handbook of Enology. The microbiology of wine and vinifications. Wiley, London, UK
Richard G, Yu S, Monsan P, Remaud-Simeon M, Morel S (2005) A novel family of glucosyl 1, 5-anhydro-D-fructose derivatives synthesised by transglucosylation with dextransucrase from Leuconostoc mesenteroides NRRL B-512 F. Carbohydr Res 340:395–401
Romaní A, Yáñez R, Garrote G, Alonso JL (2008) SSF production of lactic acid from cellulosic biosludges. Bioresour Technol 99:4247–4254
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16:944–945
Saeed AL, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J (2003) TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374–378
Sánchez-Maldonado A, Schieber A, M G (2011) Structure-function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J Appl Microbiol 111:1176–1184
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689
Siezen RJ, Tzeneva VA, Castioni A, Wels M, Phan HT, Rademaker JL, Starrenburg MJ, Kleerebezem M, Molenaar D, Van Hylckama Vlieg, Johan ET (2010) Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12:758–773
Siezen RJ, Francke C, Renckens B, Boekhorst J, Wels M, Kleerebezem M, van Hijum SA (2012) Complete resequencing and reannotation of the Lactobacillus plantarum WCFS1 genome. J Bacteriol 194:195–196
Silhavy K, Mandl K (2006) Acetobacter tropicalis in spontaneously fermented wines with vinegar fermentation in Austria. Mitt Klosterneuburg 56:102–107
Silva M, Malcata F (2000) Effect of time and temperature of fermentation on the microflora of grape pomace. Bioprocess Eng 23:17–24
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol İ (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–1123
Smokvina T, Wels M, Polka J, Chervaux C, Brisse S, Boekhorst J, van Hylckama Vlieg, Johan ET, Siezen RJ (2013) Lactobacillus paracasei comparative genomics: towards species pan-genome definition and exploitation of diversity. PLoS ONE 8:e68731
Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964
Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P (2009) The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956
Torriani S, Felis GE, Dellaglio F (2001) Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl Environ Microbiol 67:3450–3454
Treu L, Toniolo C, Nadai C, Sardu A, Giacomini A, Corich V, Campanaro S (2013) The impact of genomic variability on gene expression in environmental Saccharomyces cerevisiae strains. Environ Microbiol. doi:10.1111/1462-2920.12327
Treu L, Vendramin V, Bovo B, Giacomini A, Corich V, Campanaro S (2014) Genome sequence of Lactobacillus fabifermentans strain T30PCM01, isolated from fermenting grape marc. Genome Announc 2:e00060–14
Valera MJ, Laich F, González SS, Torija MJ, Mateo E, Mas A (2011) Diversity of acetic acid bacteria present in healthy grapes from the Canary Islands. Int J Food Microbiol 151:105–112
Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS (2005) BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 33:W455–W459
van Heel AJ, de Jong A, Montalbán-López M, Kok J, Kuipers OP (2013) BAGEL3: automated identification of genes encoding bacteriocins and (non-) bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–W453
van Nimwegen E (2003) Scaling laws in the functional content of genomes. Trends Genet 19:479–484
Vernikos GS, Parkhill J (2006) Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22:2196–2203
Versini G, Margheri G (1979) Report on the volatile constituents of grappa and organoleptic characteristics. Wines Italy 21:269–277
Wang L, Zhao B, Liu B, Yang C, Yu B, Li Q, Ma C, Xu P, Ma Y (2010) Efficient production of L-lactic acid from cassava powder by Lactobacillus rhamnosus. Bioresour Technol 101:7895–7901
Weinberg Z, Ashbell G, Azrieli A (1988) The effect of applying lactic bacteria at ensilage on the chemical and microbiological composition of vetch, wheat and alfalfa silages. J Appl Microbiol 64:1–7
Whitman WB, Goodfellow M, Kämpfer P, Busse H, Trujillo ME, Ludwig W, Suzuki K, Parte A (2012) Bergey’s manual® of systematic bacteriology. Springer, New York, NY
Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA (2008) DBD-taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res 36:D88–D92
Yamada Y, Yukphan P (2008) Genera and species in acetic acid bacteria. Int J Food Microbiol 125:15–24
Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829
Zhou F, Xu Y (2010) cBar: a computer program to distinguish plasmid-derived from chromosome-derived sequence fragments in metagenomics data. Bioinformatics 26:2051–2052
Acknowledgments
Genome sequence and 16S rDNA sequencing were performed at the UNSW Ramaciotti Centre for Gene Function Analysis (UNSW, Sydney, AU); the authors gratefully acknowledge the assistance of the staff in processing our samples. Sample preparation for scanning electron microscopy and sample visualization were performed respectively at the electron microscope facility of the Department of Biology (SME) (http://dept.bio.unipd.it/servizioME/) (University of Padova, Padua, Italy) and at the university center for scientific instruments (CUGAS) (http://www.unipd.it/cugas/) (University of Padova, Padua, Italy). Optical microscopy visualization was performed at the graphics, optical, and confocal microscopy service of the Department of Biology (University of Padova, Padua, Italy) (http://www.biologia.unipd.it/dipartimento/organizzazione/sezione-tecnico-scientifica/servizio-di-grafica-e-microscopia-ottica-e-confocale/). We express thanks to Riccardo Rosselli of the Department of Biology (University of Padua) for suggestions on metagenomic sample preparation. This study was supported, in part, by POR Veneto “Fondo Europeo di Sviluppo Regionale” 2007–2013—Asse 1, Azione 1.1.2 and by “Ricerca Scientifica fondi quota EX 60 % 2013” code 60A06-0375/13.
Author information
Authors and Affiliations
Corresponding author
Additional information
Stefano Campanaro and Laura Treu contributed equally to the work.
Rights and permissions
About this article
Cite this article
Campanaro, S., Treu, L., Vendramin, V. et al. Metagenomic analysis of the microbial community in fermented grape marc reveals that Lactobacillus fabifermentans is one of the dominant species: insights into its genome structure. Appl Microbiol Biotechnol 98, 6015–6037 (2014). https://doi.org/10.1007/s00253-014-5795-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5795-3