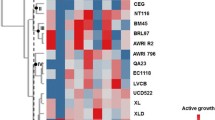
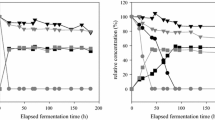
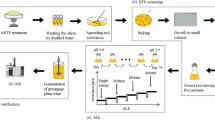
Abstract
In an era of economic globalization, the competition among wine businesses is likely to get tougher. Biotechnological innovation permeates the entire world and intensifies the severity of the competition of the wine industry. Moreover, modern consumers preferred individualized, tailored, and healthy and top quality wine products. Consequently, these two facts induce large gaps between wine production and wine consumption. Market-orientated yeast strains are presently being selected or developed for enhancing the core competitiveness of wine enterprises. Reasonable biological acidity is critical to warrant a high-quality wine. Many wild-type acidity adjustment yeast strains have been selected all over the world. Moreover, mutation breeding, metabolic engineering, genetic engineering, and protoplast fusion methods are used to construct new acidity adjustment yeast strains to meet the demands of the market. In this paper, strategies and concepts for strain selection or improvement methods were discussed, and many examples based upon selected studies involving acidity adjustment yeast strains were reviewed. Furthermore, the development of acidity adjustment yeast strains with minimized resource inputs, improved fermentation, and enological capabilities for an environmentally friendly production of healthy, top quality wine is presented.


Similar content being viewed by others
References
Agouridis N, Bekatorou A, Nigam P, Kanellaki M (2005) Malolactic fermentation in wine with Lactobacillus casei cells immobilized on delignified cellulosic material. J Agric Food Chem 53:2546–2551
Agouridis N, Kopsahelis N, Plessas S, Koutinas AA, Kanellaki M (2008) Oenococcus oeni cells immobilized on delignified cellulosic material for malolactic fermentation of wine. Bioresour Technol 99:9017–9020
Alcaide-Hidalgo JM, Moreno-Arribas MV, Martín-Álvarez PJ, Polo MC (2007) Influence of malolactic fermentation, postfermentative treatments and ageing with lees on nitrogen compounds of red wines. Food Chem 103:572–581
Aliverdieva DA, Mamaev DV, Bondarenko DI, Sholtz KF (2006) Properties of yeast Saccharomyces cerevisiae plasma membrane dicarboxylate transporter. Biochemistry (Moscow) 71:1161–1169
Ansanay V, Dequin S, Blondin B, Barre P (1993) Cloning, sequence and expression of the gene encoding the malolactic enzyme from Lactococcus lactis. FEBS Lett 332:74–80
Ansanay V, Dequin S, Camarasa C, Schaeffer V, Grivet JP, Blondin B, Salmon JM, Barre P (1996) Malolactic fermentation by engineered Saccharomyces cerevisiae as compared with engineered Schizosaccharomyces pombe. Yeast 12:215–225
Asano T, Kurose N, Tarumi S (2001) Isolation of high-malate-producing sake yeasts from low-maltose-assimilating mutants. J Biosci Bioeng 92:429–433
Baranowski K, Radler F (1984) The glucose-dependent transport of l-malate in Zygosaccharomyces bailii. Antonie Van Leeuwenhoek 50:329–340
Barre P, Dequin S (1992) Souches de levure exprimant le gene de la ldh lactique, et vecteurs utilisables pour l'Obtention desdites souches. Eur Patent 0,651,784B1
Bartowsky EJ (2005) Oenococcus oeni and malolactic fermentation-moving into the molecular arena. Aust J Grape Wine Res 11:174–187
Bartowsky EJ (2009) Bacterial spoilage of wine and approaches to minimize it. Lett Appl Microbiol 48:149–156
Bauer J, Luttik MA, Flores CL, van Dijken JP, Pronk JT, Niederberger P (1999) By-product formation during exposure of respiring Saccharomyces cerevisiae cultures to excess glucose is not caused by a limited capacity of pyruvate carboxylase. FEMS Microbiol Lett 179:107–113
Bauer R, Volschenk H, Dicks LM (2005) Cloning and expression of the malolactic gene of Pediococcus damnosus NCFB1832 in Saccharomyces cerevisiae. J Biotechnol 118:353–362
Beelman RB, Gallander JF (1979) Wine deacidification. Adv Food Res 25:1–53
Bely M, Stoeckle P, Masneuf-Pomarède I, Dubourdieu D (2008) Impact of mixed Torulaspora delbrueckii–Saccharomyces cerevisiae culture on high-sugar fermentation. Int J Food Microbiol 122:312–320
Benda I, Schmitt A (1969) Acid reduction in must by various strains of the genus Schizoaccharomyces. Weinberg Keller 16:71–83
Benito S, Palomero F, Morata A, Calderón F, Suárez-Lepe JA (2012) New applications for Schizosaccharomyces pombe in the alcoholic fermentation of red wines. Int J Food Sci Tech 47:2101–2108
Benito S, Gálvez L, Palomero F, Calderón F, Morata A, Palmero D, Suárez-Lepe JA (2013a) Schizosaccharomyces selective differential media. Afr J Microbiol Res 7:3026–3036
Benito S, Palomero F, Morata A, Calderón F, Palmero D, Suárez-Lepe JA (2013b) Physiological features of Schizosaccharomyces pombe of interest in making of white wines. Eur Food Res Technol 236:29–36
Blondin B, Dequin S (1998) Yeast, wine and genetic engineering. Biofutur 182:16–20
Bony M, Bidart F, Camarasa C, Ansanay V, Dulau L, Barre P, Dequin S (1997) Metabolic analysis of S. cerevisiae strains engineered for malolactic fermentation. FEBS Lett 410:452–456
Branduardi P, Sauer M, De Gioia L, Zampella G, Valli M, Mattanovich D, Porro D (2006) Lactate production yield from engineered yeasts is dependent from the host background, the lactate dehydrogenase source and the lactate export. Microb Cell Fact 5:4
Camarasa C, Bidard F, Bony M, Barre P, Dequin S (2001) Characterization of Schizosaccharomyces pombe malate permease by expression in Saccharomyces cerevisiae. Appl Environ Microbiol 67:4144–4151
Camarasa C, Grivet JP, Dequin S (2003) Investigation by 13C NMR and tricarboxylic acid (TCA) deletion mutant analysis of pathways for succinate formation in Saccharomyces cerevisiae during anaerobic fermentation. Microbiology 149:2669–2678
Carre E, Lafon-Lafourcade S, Bertrand A (1983) Desacidification biologiques des vins blancs secs par fermentation de I'acide malique par les levures. Connaiss Vigne Vin 17:43–53
Casas E (1999) Microorganismos responsables de alteraciones en alimentos altamente azucarados. Dissertation, Universidad Complutense de Madrid
Charpentier C, Feuillat M, Gerbaux V, Auther R (1985) La déacidification biologique des vins blancs par les Schizosaccharomyces. CR Acad Agric de France 71:425–432
Churdchai C, Haritchanan S (2000) The study of protoplast fusion between Leuconostoc oenos (malolactic fermentation bacteria) and Saccharomyces cerevisiae for industrial wine production. Proc. AABE Biennial Conference; Proc. 4th Int Conf food Sci Tech pp 121–136
Ciani M (1995) Continuous deacidification of wine by immobilized Schizosaccharomyces pombe cells: evaluation of malic acid degradation rate and analytical profiles. J Appl Microbiol 79:631–634
Ciani M, Beco L, Comitini F (2006) Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int J Food Microbiol 108:239–245
Ciani M, Comitini F, Mannazzu I, Domizio P (2010) Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res 10:123–133
Clemente-Jimenez JM, Mingorance-Cazorla L, Martínez-Rodríguez S, Las Heras-Vázquez FJ, Rodríguez-Vico F (2004) Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol 21:149–155
Colombié S, Sablayrolles JM (2004) Nicotinic acid controls lactate production by K1-LDH: a Saccharomyces cerevisiae strain expressing a bacterial LDH gene. J Ind Microbiol Biotechnol 31:209–215
Coloretti F, Zambonelli C, Castellari L, Tini V, Rainieri S (2002) The effect of dl-malic acid on the metabolism of L-malic acid during wine alcoholic fermentation. Food Technol Biotech 40:317–320
Côrte-Real M, Leão C (1990) Transport of malic acid and other dicarboxylic acids in the yeast Hansenula anomala. Appl Environ Microbiol 56:1109–1113
Côrte-Real M, Leão C (1992) Deacidification of grape juice with derepressed mutants of the yeast Hansenula anomala. Appl Microbiol Biotechnol 36:663–666
Côrte-Real M, Leão C, van Uden N (1989) Transport of L(−)malic acid and other dicarboxylic acids in the yeast Candida sphaerica. Appl Environ Microbiol 31:551–555
De Fátima M, Centeno F, Palacios A (2007) Desacidificacion Biologica de mosto a traves de la inoculacion de levadura Schizosaccharomyces pombe encapsulada como alternativa a la no produccion de aminas biogenas. International Symposium of Microbiology and Food Safety in wine “Microsafetywine”. Villafranca del Penedes, Spain, pp 20–21
Delcourt F, Taillandier P, Vidal F, Strehaiano P (1995) Influence of pH, malic acid and glucose concentrations on malic acid consumption by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 43:321–324
Denayrolles M, Aigle M, Lonvaud-Funel A (1995) Functional expression in Saccharomyces cerevisiae of the Lactococcus lactis mleS gene encoding the malolactic enzyme. FEMS Microbiol Lett 125:37–43
Dequin S (2001) The potential of genetic engineering for improving brewing, wine-making and baking yeasts. Appl Microbiol Biotechnol 56:577–588
Dequin S, Barre P (1994) Mixed lactic acid-alcoholic fermentation by Saccharomyces cerevisiae expressing the Lactobacillus casei L(+)-LDH. Biotechnology (N Y) 12:173–177
Dequin S, Baptista E, Barre P (1999) Acidification of grape musts by Saccharomyces cerevisiae wine yeast strains genetically engineered to produce lactic acid. Am J Enol Vitic 50:45–50
Dequin S, Salmon JM, Nguyen HV, Blondin B (2003) Wine yeasts. In: Boekhout T, Robert B (eds) Yeasts in food, beneficial and detrimental aspects. Berhr's, Hamburg, pp 389–412
du Toit M, Engelbrecht L, Lerm E, Krieger-Weber S (2011) Lactobacillus: the next generation of malolactic fermentation starter cultures—an overview. Food Bioprocess Technol 4:876–906
Erten H (2002) Relations between elevated temperatures and fermentation behaviour of Kloeckera apiculata and Saccharomyces cerevisiae associated with winemaking in mixed cultures. World J Microbiol Biotechnol 18:373–378
Ethiraj S, Suresh ER, Onkaraya H (1983) Controlled deacidification of Bangalore blue grape must with Schizosaccharomyces pombe. J Food Sci Technol 20:248–250
Fatichenti F, Farris GA, Deiana P, Ceccarelli S (1984) Malic acid production and consumption by selected of Saccharomyces cerevisiae under anaerobic and aerobic conditions. Appl Microbiol Biotechnol 19:427–429
Fleet GH (2003) Yeast interactions and wine flavor. Int J Food Microbiol 86:11–22
Fleet GH (2008) Wine yeasts for the future. FEMS Yeast Res 8:979–995
Fuck E, Radler F (1972) Äpfelsäurestoffwechsel bei Saccharomyces I. Der anaerobe Äpfelsäureabbau bei Saccharomyces cerevisiae. Arch Mikrobiol 87:149–164
Fuck E, Stärk G, Radler F (1973) Äpfelsäurestoffwechsel bei Saccharomyces II. Anreicherung und Eigenschaften eines Malatenzymes. Arch Mikrobiol 89:223–231
Gallander JF (1977) Deacidification of eastern table wines with Schizosaccharomyces pombe. Am J Enol Vitic 28:65–68
Gao C, Fleet GH (1995) Degradation of malic and tartaric acids by high density cell suspensions of wine yeasts. Food Microbiol 12:65–71
Gao NF, Wang SH, Li XG, Yang F (2000) Construction of yeast of reducing acid by intergeneric fusion between Saccharomyces bayanus and Schizosaccharomyces pombe. Chin J Biotechnol 16:718–722
Gao YR, Li DP, Gao NF (2006) Studies on breeding of wine yeast with good ability of reducing acid by single inactivated protoplast fusion technique. Journal of Chinese Institute of Food Science and Technology 6:106–110
Gong JX, Zheng HJ, Wu ZJ, Chen T, Zhao XM (2009) Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv 27:996–1005
Goto S, Yamazaki M, Yamakawa Y, Yokotsuka I (1978) Decomposition of l-malic acid in grape must by wine and wild yeasts. Hakko Kogaku Zasshi 56:133–135
Grobler J, Bauer F, Subden RE, Van Vuuren HJJ (1995) The mae1 gene of Schizosaccharomyces pombe encodes a permease for malate and other C4 dicarboxylic acids. Yeast 11:1485–1491
Guo LY, Tsay SS (1989) Maloalcoholic fermentation by immobilized Schizosaccharomyces pombe. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi 22:229–241
Hansen EH, Nissen P, Sommer P, Nielsen JC, Arneborg N (2001) The effect of oxygen on the survival of non-Saccharomyces yeasts during mixed culture fermentations of grape juice with Saccharomyces cerevisiae. J Appl Microbiol 91:541–547
Harrod CJ, Rodriguez SB, Thornton RJ (1997) Derepressed utilization of l-malic acid and succinic acid by mutants of Pachysolen tannophilus. J Ind Microbiol Biotechnol 18:379–383
He ZB (2004) Study on construction of deacidification wine yeast by protoplast fusion. Dissertation, Northwest A&F University
Heard GM, Fleet GH (1988) The effects of temperature and pH on the growth of yeast species during the fermentation of grape juice. J Appl Microbiol 65:23–28
Henick-Kling T (1993) Malolactic fermentation. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood Academic, Chur, pp 289–326
Hirasawa T, Ookubo A, Yoshikawa K, Nagahisa K, Furusawa C, Sawai H, Shimizu H (2009) Investigating the effectiveness of DNA microarray analysis for identifying the genes involved in l-lactate production by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 84:1149–1159
Hohmann S (1991) Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol 173:7963–7969
Hong SK, Lee HJ, Park HJ, Hong YA, Rhee IK, Lee WH, Choi SW, Lee OS, Park HD (2010) Degradation of malic acid in wine by immobilized Issatchenkia orientalis cells with oriental oak charcoal and alginate. Lett Appl Microbiol 50:522–529
Husnik JI, Volschenk H, Bauer J, Colavizza D, Luo ZL, van Vuuren HJJ (2006) Metabolic engineering of malolactic wine yeast. Metab Eng 8:315–323
Husnik JI, Delaquis PJ, Cliff MA, van Vuuren HJJ (2007) Functional analyses of the malolactic wine yeast ML01. Am J Enol Vitic 58:42–52
Ishida N, Saitoh S, Tokuhiro K, Nagamori E, Matsuyama T, Kitamoto K, Takahashi H (2005) Efficient production of l-lactic acid by metabolically engineered Saccharomyces cerevisiae with a genome-integrated l-lactate dehydrogenase gene. Appl Environ Microbiol 71:1964–1970
John RP, Anisha GS, Pandey A, Nampoothiri KM (2010) Genome shuffling: a new trend in improved bacterial production of lactic acid. Industrial Biotechnology 6:164–169
Kapsopoulou K, Kapaklis A, Spyropoulos H (2005) Growth and fermentation characteristics of a strain of the wine yeast Kluyveromyces thermotolerans isolated in Greece. World J Microb Biot 21:1599–1602
Kapsopoulou K, Mourtzini A, Anthoulas M, Nerantzis E (2007) Biological acidification during grape must fermentation using mixed cultures of Kluyveromyces thermotolerans and Saccharomyces cerevisiae. World J Microb Biot 23:735–739
Kim DH, Hong YA, Park HD (2008) Co-fermentation of grape must by Issatchenkia orientalis and Saccharomyces cerevisiae reduces the malic acid content in wine. Biotechnol Lett 30:1633–1638
Kishimoto M, Goto S (1995) Growth temperatures and electrophoretic karyotyping as tools for practical discrimination of Saccharomyces bayanus and Saccharomyces cerevisiae. J Gen Appl Microbiol 41:239–247
Kitagaki H, Kato T, Isogai A, Mikami S, Shimoi H (2008) Inhibition of mitochondrial fragmentation during sake brewing causes high malate production in sake yeast. J Biosci Bioeng 105:675–678
Kuczynski JT, Radler F (1982) The anaerobic metabolism of malate of Saccharomyces bailii and the partial purification and characterization of malic enzyme. Arch Microbiol 131:266–270
Kunicka-Styczyńska A (2009) Glucose, l-malic acid and pH effect on fermentation products in biological deacidification. Czech J Food Sci 27:319–322
Kunicka-Styczyńska A, Rajkowska K (2012) Phenotypic and genotypic diversity of wine yeasts used for acidic musts. World J Microbiol Biotechnol 28:1929–1940
Labarre C, Diviès C, Guzzo J (1996a) Genetic organization of the mle locus and identification of a mleR-like gene from Leuconostoc oenos. Appl Environ Microbiol 62:4493–4498
Labarre C, Guzzo J, Cavin JF, Diviès C (1996b) Cloning and characterization of the genes encoding the malolactic enzyme and the malate permease of Leuconostoc oenos. Appl Environ Microbiol 62:1274–1282
Lerm E, Engelbrecht L, du Toit M (2010) Malolactic fermentation: the ABC’s of MLF. S Afr J Enol Vitic 31:186–212
Li H, You L, Liu XQ, Liu YL (2006) Study of the fusant between Saccharomyces cerevisiae and Oenococcus oeni. J Northwest A&F Univ (Nat Sci Ed) 34:133–136
Liu XQ (2006) Study on breeding deacidification yeast strain for wine. Dissertation, Northwest A&F University
Liu YL, Li H (2009) Integrated expression of the Oenococcus oeni mleA gene in Saccharomyces cerevisiae. Sci Agri Sin 42:1372–1377
Maconi E, Manachini PL, Aragozzini F, Gennari C, Ricca GS (1984) A study of the maloalcoholic fermentation pathway in Schizosaccharomyces pombe. Biochem J 217:585–588
Magyar I, Panyik I (1989) Biological deacidification of wine with Schizosaccharomyces pombe entrapped in Ca-alginate gel. Am J Enol Viticu 40:233–240
Maicas S (2001) The use of alternative technologies to develop malolactic fermentation in wine. Appl Microbiol Biotechnol 56:35–39
Mayer K, Temperli A (1963) The metabolism of l-malate and other compounds by Schizosaccharomyces pombe. Arch Mikrobiol 46:321–328
Minard KI, McAlister-Henn L (1991) Isolation, nucleotide sequence analysis, and disruption of the MDH2 gene from Saccharomyces cerevisiae: evidence for three isozymes of yeast malate dehydrogenase. Mol Cell Biol 11:370–380
Minard KI, McAlister-Henn L (1992) Glucose-induced degradation of the MDH2 isozyme of malate dehydrogenase in yeast. J Biol Chem 267:17458–17464
Mora J, Barbas JI, Mulet A (1990) Growth of yeast species during the fermentation of musts inoculated with Kluyveromyces thermotolerans and Saccharomyces cerevisiae. Am J Enol Viticult 41:156–159
Mourgues J (1993) Utilisitation des résins échangeuses d’ions. Revue des Oenologues 69S:51–54
Munyon JR, Nagel CW (1977) Comparison of methods of deacidification of musts and wines. Am J Enol Viticu 28:79–87
Nakayama S, Tabata K, Oba T, Kusumoto K, Mitsuiki S, Kadokura T, Nakazato A (2012) Characteristics of the high malic acid production mechanism in Saccharomyces cerevisiae sake yeast strain no. 28. J Biosci Bioeng 114:281–285
Nevoigt E (2008) Progress in metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 72:379–412
Nissen P, Arneborg N (2003) Characterization of early deaths of non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch Microbiol 180:257–263
Oba T, Suenaga H, Nakayama S, Mitsuiki S, Kitagaki H, Tashiro K, Kuhara S (2011) Properties of a high malic acid-producing strains of Saccharomyces cerevisiae isolated from sake mash. Biosci Biotechnol Biochem 75:2025–2029
Osothsilp C (1987) Genetic and biochemical studies of malic acid metabolism in Schizosaccharomyces pombe. University of Guelph, Dissertation
Osothsilp C, Subden RE (1986) Malate transport in Schizosaccharomyces pombe. J Bacteriol 168:1439–1443
Ostergaard S, Olsson L, Nielsen J (2000) Metabolic engineering of Saccharomyces cerevisiae. Microbiol Mol Biol Rev 64:34–50
Palomero F, Morata A, Benito S, Calderón F, Suárez-Lepe JA (2009) New genera of yeasts for over-lees aging of red wine. Food Chem 112:432–441
Pathania N, Kanwar SS, Jhang T, Koundal KR, Sharma TR (2010) Application of different molecular techniques for deciphering genetic diversity among yeast isolates of traditional fermented food products of Western Himalayas. World J Microbiol Biotechnol 26:1539–1547
Peinado RA, Moreno JJ, Medina M, Mauricio JC (2005) Potential application of a glucose-transport-deficient mutant of Schizosaccharomyces pombe for removing gluconic acid from grape must. J Agric Food Chem 53:1017–1021
Peinado RA, Moreno JJ, Maestre O, Mauricio JC (2007) Removing gluconic acid by using different treatments with a Schizosaccharomyces pombe mutant: effect on fermentation byproducts. Food Chem 104:457–465
Peinado RA, Maestre O, Mauricio JC, Moreno JJ (2009) Use of a Schizosaccharomyces pombe mutant to reduce the content in gluconic acid of must obtained from rotten grapes. J Agric Food Chem 57:2368–2377
Peleg Y, Rokem JS, Goldberg I, Pines O (1990) Inducible overexpression of the FUM1 gene in Saccharomyces cerevisiae: localization of fumarase and efficient fumaric acid bioconversion to l-malic acid. Appl Environ Microbiol 56:2777–2783
Pilone JG, Ryan AF (1997) A New Zealand experience in yeast inoculation for acid reduction. Wine Ind J 12:83–86
Pines O, Even-Ram S, Elnathan N, Battat E, Aharonov O, Gibson D, Goldberg I (1996) The cytosolic pathway of l-malic acid synthesis in Saccharomyces cerevisiae: the role of fumarase. Appl Microbiol Biotechnol 46:393–399
Pines O, Shemesh S, Battat E, Goldberg I (1997) Overexpression of cytosolic malate dehydrogenase (MDH2) causes overproduction of specific organic acids in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 48:248–255
Portugal I, Ribeiro SC, Xavier AM, Centeno F, Strehaiano P (2011) Immobilised yeast grape must deacidification in a recycle fixed bed reactor. Int J Food Sci Tech 46:284–289
Postma E, Verduyn C, Scheffers WA, Van Dijken JP (1989) Enzymic analysis of the Crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 55:468–477
Presečki AV, Vasić-Rački Đ (2005) Production of l-malic acid by permeabilized cells of commercial Saccharomyces sp. strains. Biotechnol Lett 27:1835–1839
Pretorius IS (2000) Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16:675–729
Pretorius IS, Bauer FF (2002) Meeting the consumer challenge through genetically customized wine-yeast strains. Trends Biotechnol 20:426–432
Pretorius IS, du Toit M, van Rensburg P (2003) Designer yeasts for the fermentation industry of the 21st century. Food Technol Biotech 41:3–10
Pronk JT, Steensma HY, Van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633
Radler F (1986) Microbial biochemistry. Experientia 42:884–892
Radler F (1993) Yeasts—metabolism of organic acids. In: Fleet GH (ed) Wine microbiology and biotechnology. Harwood Academic, Chur, pp 165–182
Rainieri S, Zambonelli C, Giudici P, Castellari L (1998) Characterisation of thermotolerant Saccharomyces cerevisiae hybrids. Biotechnol Lett 20:543–547
Rankine BC (1966) Decomposition of l-malic acid by wine yeasts. J Sci Food Agr 17:312–316
Redzepovic S, Orlic S, Majdak A, Kozina B, Volschenk H, Viljoen-Bloom M (2003) Differential malic acid degradation by selected strains of Saccharomyces during alcoholic fermentation. Int J Food Microbiol 83:49–61
Rodriguez SB, Thornton RJ (1989) A malic acid dependent mutant of Schizosaccharomyces malidevorans. Arch Microbiol 152:564–566
Rodriguez SB, Thornton RJ (1990) Factors influencing the utilization of l-malate by yeasts. FEMS Microbiol Lett 60:17–22
Rosini G, Ciani M (1993) Influence of sugar type and level on malate metabolism of immobilized Schizosaccharomyces pombe cells. Am J Enol Vitic 44:113–117
Ruffner HP (1982) Metabolism of tartaric and malic acids in Vitis. Vitis 21:247–259
Saayman M, Viljoen-Bloom M (2006) The biochemistry of malic acid metabolism by wine yeasts—a review. S Afr J Enol Vitic 27:113–122
Saayman M, van Zyl WH, Viljoen-Bloom M (2006) Cloning, characterisation, and heterologous expression of the Candida utilis malic enzyme gene. Curr Genet 49:248–258
Sablayrolles JM (2009) Control of alcoholic fermentation in winemaking: current situation and prospect. Food Res Int 42:418–424
Saitoh S, Ishida N, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, Takahashi H (2005) Genetically engineered wine yeast produces a high concentration of l-lactic acid of extremely high optical purity. Appl Environ Microbiol 71:2789–2792
Salmon JM (1987) l-Malic acid permeation in resting cells of anaerobically grown Saccharomyces cerevisiae. Biochim Biophys Acta 901:30–34
Salmon JM, Vezinhet F, Barre P (1987) Anabolic role of l-malic acid in Saccharomyces cerevisiae in anerobiosis during alcoholic fermentation. FEMS Microbiol Lett 42:213–220
Santos J, Sousa MJ, Cardoso H, Inácio J, Silva S, Spencer-Martins I, Leão C (2008) Ethanol tolerance of sugar transport, and the rectification of stuck wine fermentations. Microbiology 154:422–430
Schuller D (2010) Better yeast for better wine-genetic improvement of Saccharomyces cerevisiae wine strains. In: Rai M, Koevics G (eds) Progress in mycology. Scientific Publishers, Jodhpur, pp 1–51
Schuller D, Casal M (2005) The use of genetically modified Saccharomyces cerevisiae strains in the wine industry. Appl Microbiol Biotechnol 68:292–304
Schümann C, Michlmayr H, Eder R, Del Hierro AM, Kulbe KD, Mathiesen G, Nguyen TH (2012) Heterologous expression of Oenococcus oeni malolactic enzyme in Lactobacillus plantarum for improved malolactic fermentation. AMB Express 2:19
Schümann C, Michlmayr H, del Hierro AM, Kulbe KD, Jiranek V, Eder R, Nguyen TH (2013) Malolactic enzyme from Oenococcus oeni heterologous expression in Escherichia coli and biochemical characterization. Bioengineered 4:147–152
Schwartz H, Radler F (1988) Formation of L (−) malate by Saccharomyces cerevisiae during fermentation. Appl Microbiol Biotechnol 27:553–560
Seo SH, Rhee CH, Park HD (2007) Degradation of malic acid by Issatchenkia orientalis KMBL 5774, an acidophilic yeast strain isolated from Korean grape wine pomace. J Microbiol 45:521–527
Servetas I, Berbegal C, Camacho N, Bekatorou A, Ferrer S, Nigam P, Drouza C, Koutinas AA (2013) Saccharomyces cerevisiae and Oenococcus oeni immobilized in different layers of a cellulose/starch gel composite for simultaneous alcoholic and malolactic wine fermentations. Process Biochem 48:1279–1284
Shi DJ, Zhang W, Zhang Y, Zhu YH (2005) Construction of malic acid-reducing yeast fusants and studies on characterization. Liquor Making 32:43–49
Silva S, Ramón-Portugal F, Andrade P, Abreu S, De Fátima TM, Strehaiano P (2003) Malic acid consumption by dry immobilized cells of Schizosaccharomyces pombe. Am J Enol Vitic 54:50–55
Sipiczki M, Ferenczy L (1977) Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol Gen Genet 151:77–81
Skory CD (2003) Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J Ind Microbiol Biotechnol 30:22–27
Snow PG, Gallander JF (1979) Deacidification of white table wines through partial fermentation with Schizosaccharomyces pombe. Am J Enol Vitic 30:45–48
Sousa MJ, Mota M, Leão C (1992) Transport of malic acid in the yeast Schizosaccharomyces pombe: evidence for a proton-dicarboxylate symport. Yeast 8:1025–1031
Sousa MJ, Mota M, Leão C (1995) Effects of ethanol and acetic acid on the transport of malic acid and glucose in the yeast Schizosaccharomyces pombe: implications in wine deacidification. FEMS Microbiol Lett 126:197–202
Steffan JS, McAlister-Henn L (1992) Isolation and characterization of the yeast gene encoding the MDH3 isozyme of malate dehydrogenase. J Biol Chem 267:24708–24715
Stephanopoulos G (2002) Metabolic engineering by genome shuffling. Nat Biotechnol 20:666–668
Su XD, Shi DJ, Cheng SM, Ma XY, Li YJ, Zhang W (2007) Construction of high performance malic acid-reducing yeast by protoplast fusion. Journal of Chinese Institute of Food Science and Technology 7:79–84
Suárez-Lepe JA, Palomero F, Benito S, Calderón F, Morata A (2012) Oenological versatility of Schizosaccharomyces spp. Eur Food Res Technol 235:375–383
Subden RE, Krizus A, Osothsilp C, Viljoen M, Van Vuuren HJJ (1998) Mutational analysis of malate pathways in Schizosaccharomyces pombe. Food Res Int 31:37–42
Taillandier P, Strehaiano P (1991) The role of malic acid in the metabolism of Schizosaccharomyces pombe: substrate consumption and cell growth. Appl Microbiol Biotechnol 35:541–543
Taillandier P, Riba JP, Strehaiano P (1988) Malate utilization by Schizosaccharomyces pombe. Biotechnol Lett 10:469–472
Taillandier P, Riba JP, Strehaiano P (1991) Malate degradation by Schizosaccharomyces yeasts included in alginate beads. Bioprocess Eng 7:141–144
Taillandier P, Gilis M, Strehaiano P (1995) Deacidification by Schizosaccharomyces: interactions with Saccharomyces. J Biotechnol 40:199–205
Taing O, Taing K (2007) Production of malic and succinic acids by sugar-tolerant yeast Zygosaccharomyces rouxii. Eur Food Res Technol 224:343–347
Temperli A, Künsch V, Mayer K, Busch I (1965) Reinigung und Eigenschaften der Malatdehydrogenase (decarboxylierent) aus Hefe. Biochim Biophys Acta 110:630–632
Thompson LM, Sutherland P, Steffan JS, McAlister-Henn L (1988) Gene sequence and primary structure of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. Biochem 27:8393–8400
Thornton RJ, Rodriguez SB (1996) Deacidification of red and white wines by a mutant of Schizosaccharomyces malidevorans under commercial winemaking conditions. Food Microbiol 13:475–482
Tokuhiro K, Ishida N, Nagamori E, Saitoh S, Onishi T, Kondo A, Takahashi H (2009) Double mutation of the PDC1 and ADH1 genes improves lactate production in the yeast Saccharomyces cerevisiae expressing the bovine lactate dehydrogenase gene. Appl Microbiol Biotechnol 82:883–890
Tristezza M, Lourenço A, Barata A, Brito L, Malfeito-Ferreira M, Loureiro V (2010) Susceptibility of wine spoilage yeasts and bacteria in the planktonic state and in biofilms to disinfectants. Ann Microbiol 60:549–556
Unterholzner O, Aurich M, Platter K (1988) Geschmacks und Geruchsfehler bei Rotweinen verursacht durch Schizosaccharomyces pombe L. Mitteilungen Klosterneuburg, Rebe und Wein, Obstbau und Früchteverwertung 38:66–70
Uribelarrea JL, De Queiroz JH, Pareilleux A (1997) Growth of Schizosaccharomyces pombe on glucose-malate mixtures in continuous cell-recycle cultures. Kinetics of substrate utilization. Appl Biochem Biotechnol 66:69–81
Valli M, Sauer M, Branduardi P, Borth N, Porro D, Mattanovich D (2006) Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl Environ Microbiol 72:5492–5499
Verduyn C, Zomerdijk TPL, van Dijken JP, Scheffers WA (1984) Continuous measurement of ethanol production by aerobic yeast suspensions with an enzyme electrode. Appl Microbiol Biotechnol 19:181–185
Viljoen M, Subden RE, Krizus A, van Vuuren HJJ (1994) Molecular analysis of the malic enzyme gene (mae2) of Schizosaccharomyces pombe. Yeast 10:613–624
Volschenk H, Viljoen M, Grobler J, Bauer F, Lonvaud-Funel A, Denayrolles M, Subden R, Van Vuuren HJJ (1997a) Malolactic fermentation in grape musts by a genetically engineered strain of Saccharomyces cerevisiae. Am J Enol Vitic 48:193–197
Volschenk H, Viljoen M, Grobler J, Petzold B, Bauer F, Subden RE, Young RA, Lonvaud A, Denayrolles M, van Vuuren HJJ (1997b) Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat Biotechnol 15:253–257
Volschenk H, Viljoen-Bloom M, Subden RE, Van Vuuren HJJ (2001) Malo-ethanolic fermentation in grape must by recombinant strains of Saccharomyces cerevisiae. Yeast 18:963–970
Volschenk H, van Vuuren HJJ, Viljoen-Bloom M (2003) Malo-ethanolic fermentation in Saccharomyces and Schizosaccharomyces. Curr Genet 43:379–391
Wang X, Gong CS, Tsao GT (1998) Production of l-malic acid via biocatalysis employing wild-type and respiratory-deficient yeasts. Appl Biochem Biotechnol 70–72:845–852
Wen LK, Wang LF, Wang GZ (2011) Degradation of l-malic and citric acids by Issatchenkia terricola. Food science 32:220–223
Williams SA, Hodges RA, Strike TL, Snow R, Kunkee RE (1984) Cloning the gene for the malolactic fermentation of wine from Lactobacillus delbrueckii in Escherichia coli and yeasts. Appl Environ Microbiol 47:288–293
Xu JY, Gao NF, Zhang Y, Sun XW (2008) Research on the construction of malic acid-degradation wine yeast by protoplast electrofusion method. Liquor-making science & technology 1:48–51
Yéramian N, Chaya C, Suárez Lepe JA (2007) l-(−)-malic acid production by Saccharomyces spp. during the alcoholic fermentation of wine (1). J Agric Food Chem 55:912–919
Yokotsuka K, Otaki A, Naitoh A, Tanaka H (1993) Controlled simultaneous deacidification and alcohol fermentation of a high-acid grape must using two immobilized yeasts, Schizosaccharomyces pombe and Saccharomyces cerevisiae. Am J Enol Vitic 44:371–377
Zelle RM, de Hulster E, van Winden WA, de Waard P, Dijkema C, Winkler AA, Geertman JMA, van Dijken JP, Pronk JT, van Maris AJ (2008) Malic acid production by Saccharomyces cerevisiae: engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl Environ Microbiol 74:2766–2777
Zelle RM, de Hulster E, Kloezen W, Pronk JT, van Maris AJA (2010a) Key process conditions for production of C4 dicarboxylic acids in bioreactor batch cultures of an engineered Saccharomyces cerevisiae strain. Appl Environ Microbiol 76:744–750
Zelle RM, Trueheart J, Harrison JC, Pronk JT, van Maris AJ (2010b) Phosphoenolpyruvate carboxykinase as the sole anaplerotic enzyme in Saccharomyces cerevisiae. Appl Environ Microbiol 76:5383–5389
Zelle RM, Harrison JC, Pronk JT, van Maris AJ (2011) Anaplerotic role for cytosolic malic enzyme in engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol 77:732–738
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WPC, del Cardayré SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Zhang XX, Jia HY, Wu B, Zhao DY, Li WX, Cheng SP (2009) Genetic analysis of protoplast fusant Xhhh constructed for pharmaceutical wastewater treatment. Bioresour Technol 100:1910–1914
Zhang XY, Hou XY, Liang F, Chen FS, Wang XH (2013) Surface display of malolactic enzyme from Oenococcus oeni on Saccharomyces cerevisiae. Appl Biochem Biotechnol 169:2350–2361
Zhao DY, Wu B, Zhang Y, Jia HY, Zhang XX, Cheng SP (2009) Identification of protoplast fusion strain Fhhh by randomly amplified polymorphic DNA. World J Microb Biot 25:1181–1188
Acknowledgments
The authors gratefully acknowledge the financial support of the National “Twelfth Five-Year” Plan for Science & Technology Support “Key Technology Research and Industry Demonstration of High Quality Fruit Wine” (2012BAD31B07). The authors would like to thank two anonymous reviewers for their comments and suggestions which greatly improved the original version of the article. Our thanks are due to Prof. Qiao-Chun Wang and Yin-Qiang Sui for assistance with this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Su, J., Wang, T., Wang, Y. et al. The use of lactic acid-producing, malic acid-producing, or malic acid-degrading yeast strains for acidity adjustment in the wine industry. Appl Microbiol Biotechnol 98, 2395–2413 (2014). https://doi.org/10.1007/s00253-014-5508-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-014-5508-y