Abstract
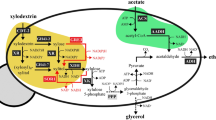
Several alcohol dehydrogenase (ADH)-related genes have been identified as enzymes for reducing levels of toxic compounds, such as, furfural and/or 5-hydroxymethylfurfural (5-HMF), in hydrolysates of pretreated lignocelluloses. To date, overexpression of these ADH genes in yeast cells have aided ethanol production from glucose or glucose/xylose mixture in the presence of furfural or 5-HMF. However, the effects of these ADH isozymes on ethanol production from xylose as a sole carbon source remain uncertain. We showed that overexpression of mutant NADH-dependent ADH1 derived from TMB3000 strain in the recombinant Saccharomyces cerevisiae, into which xylose reductase (XR) and xylitol dehydrogenase (XDH) pathway of Pichia stipitis has been introduced, improved ethanol production from xylose as a sole carbon source in the presence of 5-HMF. Enhanced furan-reducing activity is able to regenerate NAD+ to relieve redox imbalance, resulting in increased ethanol yield arising from decreased xylitol accumulation. In addition, we found that overexpression of wild-type ADH1 prevented the more severe inhibitory effects of furfural in xylose fermentation as well as overexpression of TMB3000-derived mutant. After 120 h of fermentation, the recombinant strains overexpressing wild-type and mutant ADH1 completely consumed 50 g/L xylose in the presence of 40 mM furfural and most efficiently produced ethanol (15.70 g/L and 15.24 g/L) when compared with any other test conditions. This is the first report describing the improvement of ethanol production from xylose as the sole carbon source in the presence of furan derivatives with xylose-utilizing recombinant yeast strains via the overexpression of ADH-related genes.





Similar content being viewed by others
References
Almeida JR, Röder A, Modig T, Laadan B, Lidén G, Gorwa-Grauslund MF (2008) Gorwa-Grauslund. NADH- vs. NADPH-coupled reduction of 5-hydroxymethyl furfural (HMF) and its implications on product distribution in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 78:939–945
Almeida JR, Bertilsson M, Hahn-Hägerdal B, Lidén G, Gorwa-Grauslund MF (2009) Carbon fluxes of xylose-consuming Saccharomyces cerevisiae strains are affected differently by NADH and NADPH usage in HMF reduction. Appl Microbiol Biotechnol 84:751–761
Alvira P, Tomás-Pejó E, Ballesteros M, Negro MJ (2010) Pretreatment technologies for an efficient bioethanol production process based on enzymatic hydrolysis. Bioresour Technol 101:4851–4861
Boussarsar H, Rogé B, Mathlouthi M (2009) Optimization of sugarcane bagasse conversion by hydrothermal treatment for the recovery of xylose. Bioresour Technol 100:6537–6542
Bruinenberg PM, Debot PHM, Vandijken JP, Scheffers WA (1983) The role of redox balances in the anaerobic fermentation of xylose by yeasts. Eur J appl Microbiol Biotechnol 18:287–292
Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425
Hasunuma T, Sanda T, Yamada R, Yoshimura K, Ishii J, Kondo A (2011a) Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microb Cell Fact 10:2
Hasunuma T, Sung KM, Sanda T, Yoshimura K, Matsuda F, Kondo A (2011b) Efficient fermentation of xylose to ethanol at high formic acid concentrations by metabolically engineered Saccharomyces cerevisiae. Appl Microbiol Biotechnol 90:997–1004
Ishii J, Izawa K, Matsumura S, Wakamura K, Tanino T, Tanaka T, Ogino C, Fukuda H, Kondo A (2009) A simple and immediate method for simultaneously evaluating expression level and plasmid maintenance in yeast. J Biochem 145:701–788
Katahira S, Mizuike A, Fukuda H, Kondo A (2006) Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose- and cellooligosaccharide-assimilating yeast strain. Appl Microbiol Biotechnol 72:1136–1143
Kötter P, Ciriacy M (1993) Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38:776–783
Laadan B, Almeida JR, Rådström P, Hahn-Hägerdal B, Gorwa-Grauslund M (2008) Identification of an NADH-dependent 5-hydroxymethylfurfural-reducing alcohol dehydrogenase in Saccharomyces cerevisiae. Yeast 25:191–198
Matsushika A, Watanabe S, Kodaki T, Makino K, Sawayama S (2008) Bioethanol production from xylose by recombinant Saccharomyces cerevisiae expressing xylose reductase, NADP+-dependent xylitol dehydrogenase, and xylulokinase. J Biosci Bioeng 105:296–299
Modig T, Lidén G, Taherzadeh MJ (2002) Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 363:769–776
Petersson A, Almeida JR, Modig T, Karhumaa K, Hahn-Hägerdal B, Gorwa-Grauslund MF, Lidén G (2006) A 5-hydroxymethyl furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455–464
Petschacher B, Nidetzky B (2008) Altering the coenzyme preference of xylose reductase to favor utilization of NADH enhances ethanol yield from xylose in a metabolically engineered strain of Saccharomyces cerevisiae. Microb Cell Fact 7:9
Sakamoto T, Hasunuma T, Hori Y, Yamada R, Kondo A (2012) Direct ethanol production from hemicellulosic materials of rice straw by use of an engineered yeast strain codisplaying three types of hemicellulolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. J Biotechnol 158:203–210
Sanchez B, Bautista J (1988) Effects of furfural and 5-hydroxymethylfurfural on the fermentation of Saccharomyces cerevisiae and biomass production from Candida guilliermondii. Enzyme Microb Technol 10:315–318
Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27
Tantirungkij M, Nakashima N, Seki T, Yoshida T (1993) Construction of xylose-assimilating Saccharomyces cerevisiae. J Ferment Bioeng 75:83–88
Thomsen MH, Thygesen A, Thomsen AB (2009) Identification and characterization of fermentation inhibitors formed during hydrothermal treatment and following SSF of wheat straw. Appl Microbiol Biotechnol 83:447–455
Wahlbom CF, Hahn-Hägerdal B (2002) Furfural, 5-hydroxymethyl furfural, and acetoin act as external electron acceptors during anaerobic fermentation of xylose in recombinant Saccharomyces cerevisiae. Biotechnol Bioeng 78:172–178
Watanabe S, Kodaki T, Makino K (2005) Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc. J Biol Chem 280:10340–10349
Watanabe S, Saleh AA, Pack SP, Annaluru N, Kodaki T, Makino K (2007) Ethanol production from xylose by recombinant Saccharomyces cerevisiae expressing protein engineered NADP+-dependent xylitol dehydrogenase. J Biotechnol 130:316–319
Acknowledgments
The authors would like to thank Ms. Yoshimi Hori, Ms. Shizuka Matsumura, and Ms. Hiromi Nishigaki for technical assistance. This study was supported through project P07015 of the New Energy and Industrial Technology Development Organization (NEDO), under the sponsorship of the Ministry of Economy, Trade, and Industry (METI) of Japan. This work was also supported by the Global Warming Countermeasure Technology Development Program from the Ministry of the Environment of Japan; Grants-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports and Technology (MEXT) of Japan; and a Special Coordination Fund for Promoting Science and Technology, Creation of Innovative Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe) from MEXT, Japan.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ishii, J., Yoshimura, K., Hasunuma, T. et al. Reduction of furan derivatives by overexpressing NADH-dependent Adh1 improves ethanol fermentation using xylose as sole carbon source with Saccharomyces cerevisiae harboring XR–XDH pathway. Appl Microbiol Biotechnol 97, 2597–2607 (2013). https://doi.org/10.1007/s00253-012-4376-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4376-6