Abstract
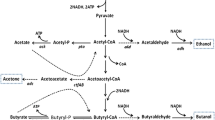
The transformation of trinitrotoluene (TNT) by several mutant strains of Clostridium acetobutylicum has been examined to analyze the maximal rate of initial transformation, determine the effects of metabolic mutations of the host on transformation rate, and to assess the cell metabolic changes brought about during TNT transformation. Little difference in the maximal rate of TNT degradation in early acid phase cultures was found between the parental ATCC 824 strain and strains altered in the acid forming pathways (phosphotransacetylase, or butyrate kinase) or in a high-solvent-producing strain (mutant B). This result is in agreement with the previous findings of a similar degradation rate in a degenerate strain (M5) that had lost the ability to produce solvent. A series of antisense constructs were made that reduced the expression of hydA, encoding the Fe-hydrogenase, or hydE and hydF, genes encoding hydrogenase maturating proteins. While the antisense hydA strain had only ∼30 % of the activity of wild type, the antisense hydE strain exhibited a TNT degradation rate around 70 % that of the parent. Overexpression of hydA modestly increased the TNT degradation rate in acid phase cells, suggesting the amount of reductant flowing into hydrogenase rather than the hydrogenase level itself was a limiting factor in many situations. The redox potential, hydrogen evolution, and organic acid metabolites produced during rapid TNT transformation in early log phase cultures were measured. The redox potential of the acid-producing culture decreased from −370 to −200 mV immediately after addition of TNT and the hydrogen evolution rate decreased, lowering the hydrogen to carbon dioxide ratio from 1.4 to around 1.1 for 15 min. During the time of TNT transformation, the treated acidogenic cells produced less acetate and more butyrate. The results show that during TNT transformation, the cells shift metabolism away from hydrogen formation to reduction of TNT and the resulting effects on cell redox cofactors generate a higher proportion of butyrate.





Similar content being viewed by others
References
Ahmad F, Hughes JB (2000) Anaerobic transformation of TNT by Clostridium. In: Spain JC, Hughes JB, Knackmuss H-J (eds) Biodegradation of nitroaromatic compounds and explosives. Lewis Publishers/CRC Press, Boca Raton, pp 185–212
Cai X, Bennett GN (2011) Improving the Clostridium acetobutylicum butanol fermentation by engineering the strain for co-production of riboflavin. J Ind Microbiol Biotechnol 38(8):1013–1025
Clark SW, Bennett GN, Rudolph FB (1989) Isolation and characterization of mutants of Clostridium acetobutylicum ATCC 824 deficient in acetoacetyl-coenzyme A:acetate/butyrate:coenzyme A transferase (EC 2.8.3.9) and in other solvent pathway enzymes. App Environ Microbial 55:970–976
Daun G, Lenke H, Reuss M, Knackmuss HJ (1998) Biological treatment of TNT-contaminated soil. 1. Anaerobic cometabolic reduction and interaction of TNT and metabolites with soil components. Environ Sci Technol 32:1956–1963
Eyers L, Smoot JC, Smoot LM, Bugli C, Urakawa H, McMurry Z, Siripong S, El-Fantroussi S, Lambert P, Agathos SN, Stahl DA (2006) Discrimination of shifts in a soil microbial community associated with TNT-contamination using a functional ANOVA of 16S rRNA hybridized to oligonucleotide microarrays. Environ Sci Technol 40(19):5867–5873
Frische T, Hoper H (2003) Soil microbial parameters and luminescent bacteria assays as indicators for in situ bioremediation of TNT-contaminated soils. Chemos 50(3):415–427
Girbal L, Soucaille P (1994) Regulation of Clostridium acetobutylicum metabolism as revealed by mixed-substrate steady-state continuous cultures: role of NADH/NAD ratio and ATP pool. J Bacteriol 176(21):6433–6438
Green EM, Boynton ZL, Harris LM, Rudolph FB, Papoutsakis ET, Bennett GN (1996) Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142(Pt 8):2079–2086
Harris LM, Welker NE, Papoutsakis ET (2002) Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol 184(13):3586–3597
Hughes JB, Wang CY, Bhadra R, Richardson A, Bennett GN, Rudolph F (1998a) Reduction of 2,4,6-trinitrotoluene by Clostridium acetobutylicum through hydroxylamino intermediates. Environ Toxicol Chem 17:343–348
Hughes JB, Wang CY, Yesland K, Richardson A, Bhadra R, Bennett GN, Rudolph F (1998b) Bamberger rearrangement during TNT metabolism by Clostridium acetobutylicum. Environ Sci Technol 32:494–500
Husemann MH, Papoutsakis ET (1990) Effects of propionate and acetate additions on solvent production in batch cultures of Clostridium acetobutylicum. Appl Environ Microbial 56(5):1497–1500
Jenkins TF, Walsh ME (1992) Development of field screening methods for TNT, 2,4-DNT, and RDX in soil. Talanta 39(4):419–428
Khan TA, Bhadra R, Hughes J (1997) Anaerobic transformation of 2,4,6-TNT and related nitroaromatic compounds by Clostridium acetobutylicum. J Ind Microbiol Biotechnol 18:198–203
King PW, Posewitz MC, Ghirardi ML, Seibert M (2006) Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system. J Bacteriol 188(6):2163–2172
Kutty R, Bennett GN (2006) Studies on inhibition of transformation of 2,4,6-trinitrotoluene catalyzed by Fe-only hydrogenase from Clostridium acetobutylicum. J Ind Microbiol Biotechnol 33(5):368–376
McGlynn SE, Ruebush SS, Naumov A, Nagy LE, Dubini A, King PW, Broderick JB, Posewitz MC, Peters JW (2007) In vitro activation of [FeFe] hydrogenase: new insights into hydrogenase maturation. J Biol Inorg Chem 12(4):443–447
Mermelstein LD, Papoutsakis ET (1993) In vivo methylation in Escherichia coli by the Bacillus subtilis phage ϕ3T methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbial 59:1077–1081
Nair RV, Green EM, Watson DE, Bennett GN, Papoutsakis ET (1999) Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J Bacteriol 181:319–330
Padda RS, Wang CY, Hughes JB, Bennett GN (2000) Mutagenicity of trinitrotoluene and its metabolites formed during anaerobic degradation by Clostridium acetobutylicum ATCC 824. Environ Toxicol Chem 19:2871–2875
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Rao G, Mutharasan R (1987) Altered electron flow in continuous cultures of Clostridium acetobutylicum induced by viologen dyes. Appl Environ Microbiol 53(6):1232–1235
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Scotcher MC, Bennett GN (2005) SpoIIE regulates sporulation but does not directly affect solventogenesis in Clostridium acetobutylicum ATCC824. J Bacteriol 187:1930–1936
Shin CY, Crawford DL (1995) Biodegradation of trinitrotoluene (TNT) by a strain of Clostridium bifermentans. In: Hinchee RW, Fredrickson J, Alleman BC (eds) Bioaugmentation for site remediation. Battelle Press, Columbus
Siciliano SD, Gong P, Sunahara GI, Greer CW (2000) Assessment of 2,4,6-trinitrotoluene toxicity in field soils by pollution-induced community tolerance, denaturing gel electrophoresis, and seed germination assay. Environ Toxicol Chem 19:2154–2160
Spain JC (1995) Biodegradation of nitroaromatic compounds. Annu Rev Microbiol 49:634–649
Stim-Herndon KP, Nair R, Papoutsakis ET, Bennett GN (1996) Analysis of degenerate variants of Clostridium acetobutylicum ATCC 824. Anaerobe 2:11–18
Tan EL, Ho CH, Griest WH, Tyndall RL (1992) Mutagenicity of trinitrotoluene and its metabolites formed during composting. J Toxicol Environ Health 36:165–172
Tummala SB, Welker NE, Papoutsakis ET (2003) Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. J Bacteriol 185(6):1923–1934
Vasconcelos I, Girbal L, Soucaille P (1994) Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol 176(5):1443–1450
Wang J, Zhu J, Bennett GN, San KY (2011) Succinate production from different carbon sources under anaerobic conditions by metabolic engineered Escherichia coli strains. Methods Eng 13(3):328–335
Watrous MM, Clark S, Kutty R, Huang S, Rudolph FB, Hughes JB, Bennett GN (2003) 2,4,6-trinitrotoluene reduction by an Fe-only hydrogenase in Clostridium acetobutylicum. Appl Environ Microbiol 69(3):1542–1547
Won WD, Salvo LHd, Ng J (1976) Toxicity and mutagenicity of 2,4,6-trinitrotoluene and its microbial metabolites. Appl Environ Microbiol 31:576–580
Wu Y, Luo Y, Zou D, Ni J, Liu W, Teng Y, Li Z (2008) Bioremediation of polycyclic aromatic hydrocarbons contaminated soil with Monilinia sp.: degradation and microbial community analysis. Biodegradation 19(2):247–257
Zhang C, Bennett GN (2005) Biodegradation of xenobiotics by anaerobic bacteria. Appl Microbial Biotechnol 67:600–618
Zhao Y, Hindorff LA, Chuang A, Monroe-Augustus M, Lyristis M, Harrison ML, Rudolph FB, Bennett GN (2003) Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 69(5):2831–2841
Zhao Y, Tomas CA, Rudolph FB, Papoutsakis ET, Bennett GN (2005) Intracellular butyryl phosphate and acetyl phosphate concentrations in Clostridium acetobutylicum and their implications for solvent formation. Appl Environ Microbiol 71(1):530–537
Acknowledgments
This project was supported by the Army Research Office, grant number W911NF0910119. Also, we would like to thank Mary Harrison and David French for assistance with this research and data management.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cai, X., Servinsky, M., Kiel, J. et al. Analysis of redox responses during TNT transformation by Clostridium acetobutylicum ATCC 824 and mutants exhibiting altered metabolism. Appl Microbiol Biotechnol 97, 4651–4663 (2013). https://doi.org/10.1007/s00253-012-4253-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-4253-3