Abstract
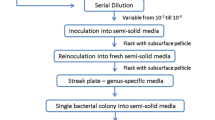
Nineteen different steroid-degrading bacteria were isolated from soil samples by using selective media containing either cholesterol or deoxycholate as sole carbon source. Strains that assimilated cholesterol (17 COL strains) were gram-positive, belonging to the genera Gordonia, Tsukamurella, and Rhodococcus, and grew on media containing other steroids but were unable to use deoxycholate as sole carbon source. Surprisingly, some of the COL strains unable to grow using deoxycholate as sole carbon source were able to catabolize other bile salts (e.g., cholate). Conversely, strains able to grow using deoxycholate as the sole carbon source (two DOC isolates) were gram-negative, belonging to the genus Pseudomonas, and were unable to catabolize cholesterol and other sterols. COL and DOC were included into the corresponding taxonomic groups based on their morphology (cells and colonies), metabolic properties (kind of substrates that support bacterial growth), and genetic sequences (16S rDNA and rpoB). Additionally, different DOC21 Tn5 insertion mutants have been obtained. These mutants have been classified into two different groups: (1) those affected in the catabolism of bile salts but that, as wild type, can grow in other steroids and (2) those unable to grow in media containing any of the steroids tested. The identification of the insertion point of Tn5 in one of the mutants belonging to the second group (DOC21 Mut1) revealed that the gene knocked-out encodes an A-ring meta-cleavage dioxygenase needed for steroid catabolism.




Similar content being viewed by others
References
Adékambi T, Drancourt M, Raoult D (2009) The rpoB gene as a tool for clinical microbiologists. Trends Microbiol 17:37–45
Anzai Y, Kim H, Park J-Y, Wakabayashi H, Oyaizu H (2000) Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50:1563–1589
Arcos M, Olivera ER, Arias S, Naharro G, Luengo JM (2010) The 3, 4-dihydroxyphenylacetic acid catabolon, a catabolic unit for degradation of biogenic amines tyramine and dopamine in Pseudomonas putida U. Environ Microbiol 12:1684–1704
Bagdasarian M, Lurz R, Ruckert B, Franklin FC, Bagdasarian MM, Frey J, Timmis KN (1981) Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247
Birkenmaier A, Holert J, Erdbrink H, Moeller HM, Friemel A, Schoenenberger R, Suter MJ, Klebensberger J, Philipp B (2007) Biochemical and genetic investigation of initial reactions in aerobic degradation of the bile acid cholate in Pseudomonas sp. strain Chol1. J Bacteriol 189:7165–7173
Björkhem I, Eggertsen G (2001) Genes involved in initial steps of bile acid synthesis. Curr Opin Lipidol 12:97–103
Campanella L, Favero G, Mastrofini D, Tomassetti M (1996) Toxicity order of cholanic acids using an immobilised cell biosensor. J Pharm Biomed Anal 14:1007–1013
Colborn T (2004) Endocrine disruption overview: are males at risk? Adv Exp Med Biol 545:189–201
Drzyzga O, Navarro Llorens JM, Fernández de las Heras L, García Fernández E, Perera J (2009) Gordonia cholesterolivorans sp. nov., a cholesterol-degrading actinomycete isolated from sewage sludge. Int J Syst Evol Microbiol 59:1011–1015
Fahrbach M, Kuever J, Meinke R, Kämpfer P, Hollender J (2006) Denitrasoma oestradiolicum gen. nov., sp. nov., a 17β-oestradiol-degrading, denitrifying betaproteobacterium. Int J Syst Evol Microbiol 56:1547–1552
Fahrbach M, Kuever J, Remesch M, Huber BE, Kämpfer P, Dott W, Hollender J (2008) Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int J Syst Evol Microbiol 58:2215–2223
Fernández de las Heras L, García Fernández E, Navarro Llorens JM, Perera J, Drzyzga O (2009) Morphological, physiological, and molecular characterization of a newly isolated steroid-degrading Actinomycete, identified as Rhodococcus ruber strain Chol-4. Curr Microbiol 59:548–553
Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) (1994) Methods for general and molecular bacteriology. American Society for Microbiology, Washington
Gibson DT, Koch JR, Kallio RE (1968) Oxidative degradation of aromatic hydrocarbons by microorganisms. I. Enzymatic formation of catechol from benzene. Biochemistry 7:2653–2662
Grant SG, Jessee J, Bloom FR, Hanahan D (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation–restriction mutants. Proc Natl Acad Sci USA 87:4645–4649
Gunn JS (2000) Mechanisms of bacterial resistance and response to bile. Microbes Infect 2:907–913
Hayakawa S (1982) Microbial transformation of bile acids. A unified scheme for bile acid degradation, and hydroxylation of bile acids. Z Allg Mikrobiol 22:309–326
Herrero M, de Lorenzo V, Timmis KN (1990) Transposon vector containing non-antibiotic markers for cloning and stable chromosomal insertion of foreign DNA in Gram-negative bacteria. J Bacteriol 172:6557–6567
Holt JG, Krieg NR, Sneath PHA, Stalely JT, Williams ST (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams & Wilkins, Baltimore
Horinouchi M, Yamamoto T, Taguchi K, Arai H, Kudo T (2001) Meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441. Microbiology 147:3367–3375
Horinouchi M, Hayashi T, Koshino H, Kurita T, Kudo T (2005) Identification of 9, 17-dioxo-1, 2, 3, 4, 10, 19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2, 4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441. Appl Environ Microbiol 71:5275–5281
Horinouchi M, Kurita T, Hayashi T, Kudo T (2010) Steroid degradation genes in Comamonas testosteroni TA441: isolation of genes encoding a Δ4(5)-isomerase and 3α- and 3β-dehydrogenases and evidence for a 100 kb steroid degradation gene hot spot. J Steroid Biochem Mol Biol 122:253–263
Kim D, Lee JS, Kim J, Kang SJ, Yoon JH, Kim WG, Lee CH (2007) Biosynthesis of bile acids in a variety of marine bacterial taxa. J Microbiol Biotechnol 17:403–407
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 44:301–307
Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MH, Roop RM, Peterson KM (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176
Kurdi P, Kawanishi K, Mizutani K, Yokota A (2006) Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol 188:1979–1986
Kurisu F, Ogura M, Saitoh S, Yamazoe A, Yagi O (2010) Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp. J Biosci Bioeng 109:576–582
Lamb DC, Jackson CJ, Warrilow AGS, Maning NJ, Kelly DE, Kelly SL (2007) Lanosterol biosynthesis in the prokaryote Methylococcus capsulatus: insight into the evolution of sterol biosynthesis. Mol Biol Evol 24:1714–1721
Länge R, Hutchinson TH, Croudace CP, Siegmund F, Schweinfurth H, Hampe P, Panter GH, Sumpter JP (2001) Effects of the synthetic estrogen 17α-ethinylestradiol on the life-cycle of the fathead minnow (Pimephales promelas). Environ Toxicol Chem 20:1216–1227
Lange A, Katsu Y, Ichikawa R, Paull GC, Chidgey LL, Coe TS, Iguchi T, Tyler CR (2008) Altered sexual development in roach (Rutilus rutilus) exposed to environmental concentrations of the pharmaceutical 17α-ethinylestradiol and associated expression dynamics of aromatases and estrogen receptors. Toxicol Sci 106:113–123
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948
Martínez-Blanco H, Reglero A, Rodríguez-Aparicio LB, Luengo JM (1990) Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. A specific enzyme for the catabolism of phenylacetic acid. J Biol Chem 265:7084–7090
Mathieu JM, Mohn WW, Eltis LD, LeBlanc JC, Stewart GR, Dresen C, Okamoto K, Alvarez PJ (2010) 7-Ketocholesterol catabolism by Rhodococcus jostii RHA1. Appl Environ Microbiol 76:352–355
Olivera ER, Miñambres B, García B, Muñiz C, Moreno MA, Fernández A, Díaz E, García JL, Luengo JM (1998) Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl–CoA catabolon. Proc Natl Acad Sci USA 95:6419–6424
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Petrusma M, Dijkhuizen L, van der Geize R (2009) Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl Environ Microbiol 75:5300–5307
Prieto AI, Ramos-Morales F, Casadesús J (2004) Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787–1794
Prieto AI, Ramos-Morales F, Casadesús J (2006) Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575–584
Ridlon JM, Kang DJ, Hylemon PB (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Selvaraj G, Iyer VN (1983) Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol 156:1292–1300
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetic analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tayeb L, Ageron E, Grimont F, Grimont PAD (2005) Molecular phylogeny of the genus Pseudomonas based on rpoB sequences and application for the identification of isolates. Res Microbiol 156:763–773
Uhía I, Galán B, Morales V, García JL (2011) Initial step in the catabolism of cholesterol by Mycobacterium smegmatis mc2 155. Environ Microbiol 13:943–959
Van der Geize R, Yam K, Heuser T, Wilbrink MH, Hara H, Anderton MC, Sim E, Dijkhuizen L, Davies JE, Mohn WW, Eltis LD (2007) A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci USA 104:1947–1952
Wollam J, Antebi A (2011) Sterol regulation of metabolism, homeostasis, and development. Annu Rev Biochem 80:885–916
Yam KC, D'Angelo I, Kalscheuer R, Zhu H, Wang J-X, Snieckus V, Ly LH, Converse PJ, Jacobs WR Jr, Strynadka N, Eltis LD (2009) Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog 5(3):e1000344. doi:10.1371/journal.ppat.1000344
Zhi X-Y, Li W-J, Stackebrandt E (2009) An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 59:589–608
Acknowledgments
This work has been supported by grants BFU2009-11545-C03-01 from Ministerio de Ciencia e Innovación (Spain) and LE246A11-2 from Junta de Castilla y León (Consejería de Educación, Spain). JR, AB, and EM are recipient of fellowships given by the Ministerio de Ciencia e Innovación (JR and AB) and by the Ministerio de Educación y Ciencia (EM).
Author information
Authors and Affiliations
Corresponding author
Additional information
E. Merino and A. Barrientos contributed equally to this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
(DOCX 12 kb)
Supplementary Table 2
(DOCX 12 kb)
Supplementary Table 3
(DOCX 13 kb)
Supplementary Table 4
(DOCX 27 kb)
Supplementary Table 5
(DOCX 16 kb)
Supplentary Table 6
(DOCX 12 kb)
Suppl. Figure 1
Structure of the steroid compounds tested in this work. 1. Cholesterol; 2. Ergosterol; 3. β-Sitosterol; 4. Stigmasterol; 5. 5β-Cholanate; 6. Lithocholate; 7. Chenodeoxycholate; 8. Ursodeoxycholate; 9. Deoxycholate; 10. Cholate; 11. Dehydrocholate; 12. Progesterone; 13. 5-Pregnen-3β-ol-20-one; 14. Testosterone; 15. 17α-Methyltestosterone; 16. 1-Dehydro-17α-Methyltestosterone; 17. Prednisone; 18. Androstanolone; 19. β-Estradiol; 20. trans-dehydroandrosterone; 21. Estrone; 22. trans-androsterone; 23. 4-Androstene-3,17-dione. Carbon atoms are numbered over colesterol structure (1). (JPEG 1.36 MB)
Suppl. Figure 2
Morphology of the colonies (a) and cells (b) of the COL isolates. Strains numbered 1–8 belong to the genus Gordonia: COL11 (1), COL17 (2), COL19 (3), COL20 (4), COL21 (5), COL23 (6), COL26 (7), and COL33 (8); strains numbered 9–11 belong to the genus Tsukamurella: COL14 (9), COL16 (10), and COL18 (11); and strains numbered 12–17 belong to the genus Rhodococcus: COL22 (12), COL25 (13), COL27 (14), COL28 (15). COL29 (16), and COL30 (17). (a) optical microscopy images, bar indicates 1 mm; (b) scanning microscopy images, bar indicates 1 μm. (JPEG 162 kb)
Suppl. Figure 3
(A and C) Optical microscopy images of the colonies and (B and D) scanning microscopy images of the cells of strains DOC19 (A and B) and DOC21 (C and D). In the optical microscopy images (A and C), the bar indicates 1 mm; in the scanning microscopy photographs (B and D), the bar indicates 1 μm. (JPEG 34 kb)
Rights and permissions
About this article
Cite this article
Merino, E., Barrientos, A., Rodríguez, J. et al. Isolation of cholesterol- and deoxycholate-degrading bacteria from soil samples: evidence of a common pathway. Appl Microbiol Biotechnol 97, 891–904 (2013). https://doi.org/10.1007/s00253-012-3966-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-012-3966-7