Abstract
In this study, dideoxy sequencing and 454 high-throughput sequencing were used to analyze diversities of the ammonia monooxygenase (amoA) genes and the 16S rRNA genes of ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) in six municipal wastewater treatment plants. The results showed that AOB amoA genes were quite diverse in different wastewater treatment plants while the 16S rRNA genes were relatively conserved. Based on the observed complexity of amoA and 16S rRNA genes, most of the AOB can be assigned to the Nitrosomonas genus, with Nitrosomonas ureae, Nitrosomonas oligotropha, Nitrosomonas marina, and Nitrosomonas aestuarii being the four most dominant species. From the sequences of the AOA amoA genes, most AOA observed in this study belong to the CGI.1b group, i.e., the soil lineage. The AOB amoA and 16S rRNA genes were quantified by quantitative PCR and 454 high-throughput pyrosequencing, respectively. Although the results from the two approaches show some disconcordance, they both indicated that the abundance of AOB in activated sludge was very low.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrification, the biological oxidation of ammonium to nitrite and nitrate, is an essential process in nitrogen cycling and wastewater treatment bioreactors. Several groups of microorganisms are involved in the two-step process: ammonia-oxidizing archaea (AOA), ammonia-oxidizing bacteria (AOB), and nitrite-oxidizing bacteria (NOB). In the first step, AOA and/or AOB oxidize NH3 to NO −2 , and in the second step NOB oxidize NO −2 to NO −3 . The first step is rate limiting and has been relatively well studied (Limpiyakorn et al. 2006; Park and Noguera 2004; Tokutomi et al. 2010). Although some heterotrophic bacteria (Robertson and Kuenen 1990) and anaerobic ammonia-oxidizing (anammox) bacteria (Strous et al. 1999) can also oxidize ammonia to nitrite, AOA and AOB are thought to be the main contributors for environmental ammonia oxidation and in bioreactors (Nicol and Schleper 2006). According to the previous reports, Nitrosomonas and Nitrosospira are the most important genera of AOB in activated sludge (Park and Noguera 2004; Purkhold et al. 2000). Of the two genera, Nitrosomonas has been shown to be the dominant AOB in many bioreactors (Limpiyakorn et al. 2006; Park and Noguera 2004; Wells et al. 2009). By contrast, Nitrosospira sp. were rarely found in activated sludge (Schramm et al. 1999), probably due to their relative low growth rate (Siripong and Rittmann 2007) resulting in an underrepresentation in bioreactors. The limited studies of AOA species in activated sludge showed a composition different from those existing in other environments such as soil, water column, and sediment (Park et al. 2006). Moreover, their abundances seemed to be much lower (four orders or more) than AOB based on analysis of amoA gene copy number (Jin et al. 2010; Limpiyakorn et al. 2010; Wells et al. 2009).
In this study, the AOA and AOB diversity in six bioreactors from the wastewater treatment plants (WWTPs) in four countries was investigated using amoA and 16S rRNA genes as the biomarkers. High-throughput pyrosequencing of the 16S rRNA genes was used to tabulate the AOB diversity and their relative abundance in the total bacterial community. Quantitative polymerase chain reaction (qPCR) was also used to quantify AOB amoA genes in the same activated sludge samples. Several SYBR green-based qPCR systems have been tried to quantify AOA amoA gene. However, non-specific amplification and/or the formation of primer dimer hindered the accurate quantification of AOA amoA using this approach.
Materials and methods
Activated sludge sampling and DNA extraction
In this study, activated sludge samples were taken from aeration tanks of eight full-scale WWTPs treating municipal wastewater in China, Singapore, Canada, and the USA. Relevant parameters about these WWTPs are shown in Supplementary Table S1. Sludge samples from the aeration tank were fixed with 50% ethanol (v/v) on site before transporting to the laboratory for DNA extraction. Ten milliliters of fixed activated sludge samples was centrifuged at 4,000 rpm for 10 min at 4 °C. Approximately 200 mg of sample pellet was recovered for DNA extraction in duplicate with a FastDNA® SPIN Kit for Soil (Qiagen, CA, USA), which was found to be the most suitable DNA extraction method for the samples in this study, as being compared with other commercial reagents.
PCR and quantitative PCR
Primer set amoA-1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA-2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) (Rotthauwe et al. 1997) was used to amplify bacterial amoA gene in a 30-μl mixture containing 0.2 μl of TaKaRa Ex TaqTM, 3 μl of 10× Ex Taq Buffer (TaKaRa), 3 μl of 10 mM dNTP mixture (TaKaRa), 0.2 μM of each primer, and 20–50 ng of genomic DNA. Thermal cycling parameters followed the protocol of Rotthauwe and colleagues (1997). AOB amoA gene copy numbers were quantified by using an iCycler IQ System (Bio-Rad, Hercules, CA, USA) in triplicate with primer set amoA-1F/amoA-2R. Quantitative real-time PCR amplification was performed in a total volume of 30 μl containing 15 μl of iQTM SYBR® Green Supermix (Bio-Rad), 5 μl of DNA template with the concentration of about 1 ng/μl, and 0.3 μM of each primer using the same cycling conditions. The archaeal amoA gene was amplified by PCR using the primer set Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) (Francis et al. 2005). PCR amplification was performed in a 50-μl volume comprising 25 μl 2× MightyAmp Buffer (TaKaRa), 1 μl (1.25 U) MightyAmp DNA Polymerase (TaKaRa), 0.3 μM of each primer, and 20–50 ng of genomic DNA. PCR was first incubated at 98 °C for 1 min and was followed by 35 cycles at 98 °C for 10 s, 60 °C for 15 s, and 68 °C for 60 s. The PCR products were visualized by agarose (1%) gel electrophoresis in the presence of suitable size markers.
For high-throughput 454 pyrosequencing, the bacterial DNA was amplified with a set of primers targeting the hypervariable V4 region of the 16S rRNA gene. The forward primer is 5′-AYTGGGYDTAAAGNG-3′ and the reverse primers are the mixture of four equally mixed primers: 5′-TACCRGGGTHTCTAATCC-3′, 5′-TACCAGAGTATCTAATTC-3′, 5′-CTACDSRGGTMTCTAATC-3′, and 5′-TACNVGGGTATCTAATCC-3′ (RDP's Pyrosequencing Pipeline, http://pyro.cme.msu.edu/pyro/help.jsp). Barcodes that allowed sample multiplexing during pyrosequencing were incorporated between the 454 adaptor A and the forward primer.
Dideoxy DNA sequencing
PCR products were purified by using PCRquick-spinTM PCR Product Purification Kit (iNtRON Biotechnology, Korea). The purified PCR products were ligated to pMD®18-T Vector (TaKaRa). Recombinant plasmid was transformed into E. coli and white colonies that grew on LB plates containing ampicillin (60 μg/ml), X-Gal, and IPTG were picked to conduct colony PCR amplification with the primer set M13F and M13R. The PCR products were purified and sequenced by ABI 3730xl capillary sequencers (Applied Biosystems, Foster City, CA, USA) with the primer M13F.
OTU definition and phylogenetic analysis
An AOB amoA gene clone library was constructed for each of the eight activated sludge samples (sample IDs—CN-NJ-SJ, CN-HR-UN, SG-SG-UP, CN-QD-TD, CN-SH-TS, CN-BJ-JX, US-CO-CO, and CA-GP-GP). Individual AOA amoA gene clone libraries were successfully constructed for samples CN-NJ-SJ, CN-HR-UN, and SG-SG-UP, while the other samples failed to generate library products. Approximately 20 clones in each clone library were selected randomly and sequenced. The resulting AOB amoA gene sequences were aligned and the Jukes–Cantor distances between subsequent pairs of sequences were calculated with DNADIST from the PHYLIP package (http://www.phylip.com/), and were grouped into 43 OTUs with a distance cut-off of 3%. In order to construct the phylogenetic tree, one sequence in each OTU was selected, merged, and aligned with the reference sequences from NCBI Entrez Database. The neighbor-joining phylogenetic tree of AOB amoA gene sequences was created by MEGA software (Kumar et al. 2008). The translated protein sequences were assigned to different OTUs with a 3% distance cut-off. The phylogenetic tree of AOB AmoA protein sequences was also created by MEGA software. AOA amoA gene and AOA AmoA protein sequences were analyzed in the same manner as that of AOB. The AOB 16S rRNA gene sequences were also classified into different OTUs with a 3% distance cut-off. In addition, Good’s estimator of coverage (Good 1953) was calculated for each AOA and AOB amoA gene clone library under 3% distance cut-off (Table S2).
High-throughput pyrosequencing
PCR amplicon libraries were prepared using a minimal number of amplification cycles (25 cycles) to minimize the accumulation of PCR artifacts. Amplicons were purified using PCRquick-spinTM columns (iNtRON Biotechnology). Equal amounts of amplicon products bearing individual sequence barcode for each sludge samples were combined for multiplex pyrosequencing on the Roche 454 FLX Titanium platform (Roche, Nutley, NJ, USA).
Sequence analysis and phylogenetic assignment
Following pyrosequencing, Python scripts were written to (1) remove sequences containing more than one ambiguous base (‘N’), (2) check the sequence integrity of the barcodes and partitioned the multiplex reads to the individual samples, and (3) remove sequence reads shorter than 150 bases. The resulting filtered reads were then compared with the Greengenes 16S rRNA gene database (DeSantis et al. 2006) using NCBI’s BLASTN tool (Altschul et al. 1990) with default parameters set to a maximum hit number of 100 (Claesson et al. 2009). Sequences were then assigned to NCBI taxonomies with MEGAN (Huson et al. 2007) using the Lowest Common Ancestor (LCA) algorithm and default parameters (absolute cut-off—BLAST bitscore 35; relative cut-off—10% of the top hits).
Accession numbers
The sequences obtained from clone library in this study were deposited in GenBank under accession numbers JF271927–JF271985. The pyrosequencing results are deposited into the NCBI short reads archive database (accession number SRA026842.2).
Results
Diversity of AOB amoA gene and 16S rRNA gene in different activated sludge samples
Table S2 suggested that the coverage of the AOB amoA gene clone libraries was over 70% except sample CN-SH-TS with a relatively low coverage (65%). As shown in Fig. 1, a total of 43 OTUs were generated based on 163 AOB amoA gene sequences in eight clone libraries with a 3% distance cut-off. Figure 1 reveals an interesting phenomenon that relatively few amoA OTUs were shared among the activated sludge samples, indicating that the amoA genes were quite diverse and not widely disseminated.
Figure 2 illustrated that most of these bacterial amoA OTUs were affiliated to Nitrosomonas genus with Nitrosomonas ureae, Nitrosomonas oligotropha, Nitrosomonas marina, and Nitrosomonas aestuarii being the four most dominant species. However, only two OTUs (OTU-17 and OTU-32) were in the Nitrosospira lineage and these two OTUs represented only eight sequences in the total 163 sequences, indicating the low abundance of Nitrosospira in various types of activated sludge.
Neighbor-joining phylogenetic tree based on bacterial amoA gene sequences. The evolutionary distances were computed using the Jukes–Cantor method and are in the units of the number of base substitutions per site. Bootstrap values are indicated on branch nodes. Sequences obtained in this study are shown with “OTU-” in the names. Other sequences were obtained from GenBank. The tree was out-grouped with Nitrosococcus halophilus (gammaproteobacterial AOB) amoA sequence
Figure S1 shows the relative abundance and distribution of AOB AmoA protein OTUs in different activated sludge samples. Compared with AOB amoA gene OTUs (shown in Fig. 1), the diversity of AmoA protein (a total of 26 OTUs) was less than that of amoA gene (43 OTUs) due to codon wobble positions. Twelve OTUs of AOB AmoA protein were shared by at least two activated sludge samples. Our findings indicate that a number of different amoA genotypes encode essentially the identical or very similar proteins possibly reflecting the functional constraints in efficient ammonia oxidation. As shown in the phylogenetic tree of AOB AmoA protein in Fig. S2, most OTUs were affiliated with N. ureae, N. oligotropha, N. marina, and N. aestuarii lineages, exactly the same as the amoA gene phylogenetic tree. There were only a few OTUs in Nitrosospira lineage and N. europaea lineage, which was also identical with the above amoA gene phylogenetic analysis.
In addition to the amoA gene, the 16S rRNA gene was also used to investigate the AOB community in six samples (i.e., CN-NJ-SJ, CN-QD-TD, CN-HR-UN, US-CO-CO, SG-SG-UP, and CA-GP-GP) by high-throughput pyrosequencing. As shown in Table 1, more than 20,000 16S rRNA gene fragment sequences were obtained for each sample site. These sequences were assigned to NCBI taxonomies using BLAST and MEGAN software. A tabulation of AOB-like sequences is shown in Table 1. The filtered reads were merged, aligned, and classified into 18 OTUs with a Jukes–Cantor distance cut-off of 3%. Figure 3 shows that most (about 61%) AOB 16S rRNA OTUs were shared by more than two sample sites, and two major OTUs, i.e., OTU-14 and OTU-15, accounted for 49% of total sequences. These results implied the AOB 16S rRNA genes are much more conservative than amoA gene and AmoA protein.
Abundance of AOB in activated sludge
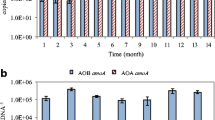
Along with direct tabulation of AOB 16S rRNA genes by pyrosequencing, we also quantify AOB abundances in the activated sludge samples by qPCR, normalizing the AOB amoA gene copy number against per nanogram of the genomic DNA extracted from activated sludge. Table 1 shows the abundances of AOB amoA genes in different activated sludge samples quantified by qPCR. Sample CN-SH-TS has the most abundant AOB amoA gene copy numbers among the eight activated sludge samples tested. Table 1 lists the total sequences obtained from pyrosequencing and the AOB-like sequences in each of the six samples. AOB sequences were successfully obtained in five out of the six samples, and the percentages range from 0.29% to 0.64%. For US-CO-CO, a sample from Columbia, USA, no AOB sequences were obtained, probably due to the low abundance of AOB in this wastewater treatment plant, in agreement with amoA gene quantification results.
Diversity of AOA amoA gene in different activated sludge samples
In the present study, multiple experiments were attempted to amplify amoA gene from the eight samples using four PCR systems (including TaKaRa Ex Taq, TaKaRa Mighty Amp DNA Polymerase, Sigma Taq DNA Polymerase, and Bio-Rad SYBR® Green Supermix) at different thermal conditions (annealing temperature from 50 to 60 °C). Amplification was successful from only five samples as shown in Fig. S3 and only three samples were eventually successfully cloned and sequenced. Additionally, the presence of primer dimers was very serious in all samples and it was not possible to quantify amoA using SYBR® Green qPCR approach applied in this study (Supporting information Fig. S3).
However, it is still of interest to have a preliminary assessment of the diversity of AOA amoA gene in the activated sludge samples by exploring the sequences obtained from the clone libraries of the samples CN-NJ-SJ, CN-HR-UN, and SG-SG-UP, all of which had Good’s estimator of coverage over 80% (Table S2). The 56 sequences from the three samples were classified into 15 OTUs using a distance cut-off of 3% (Fig. 4). Different from AOB amoA OTUs (Fig. 1), some AOA amoA OTUs were shared among multiple samples. Judging from the results in this study, it was found that the AOA amoA genes are not as diverse as AOB amoA gene in different activated sludge samples. While it should be mentioned that the sample amount in this study was kind of limited, thus this issue needs more studies to be confirmed in the future. As shown in the phylogenetic tree of AOA amoA gene sequences in Fig. 6, most of the sequences obtained in this study were distinctly different from the previously reported sequences, especially the sequences recovered from marine and sediment. Only two OTUs (OTU-10 and OTU-11) were grouped into the marine and sediment lineages.
Discussion
Diversity of AOB amoA gene and 16S rRNA gene in different activated sludge samples
A previous study reported that the amoA gene similarities among different AOB species ranged from 65% to 100% (Purkhold et al. 2000). The similarities of the OTUs obtained in this study were 62% to 100%. The high diversity of the amoA gene found in this study was likely due to the different geographic locations of these sludge samples and the different operation/sewage conditions in these WWTPs.
The results in this study were consistent with previous reports that showed Nitrosomonas instead of Nitrosospira being the dominant AOB in nitrification bioreactors in WWTP (Park and Noguera 2004; Wells et al. 2009). Notably, seven OTU-17-associated sequences were all from the sample CN-HR-UN taken from Harbin, a city in northern China. The latitude of this city is the highest among those sampled in this study. Our observation is consistent with the previous report that Nitrosospira spp. prefer low temperatures of 4–10 °C (Avrahami et al. 2003; Siripong and Rittmann 2007). We also observe a small number of OTUs belonging to Nitrosomonas europaea lineage and several OTUs (OTU-1, OTU-3, OTU-22, and OTU-26) grouped far away from the known AOB species.
It was reported that the AOB 16S rRNA gene similarities generally ranged from 90% to 100%, much higher than those of amoA gene (about 65–100%) and AmoA protein (about 72.5–100%) (Purkhold et al. 2000). The phylogenetic tree in Fig. 5 shows that all of these sequences obtained in this study were grouped in Nitrosomonas genus. There were 12 OTUs in Nitrosomonas oligotropha group, four OTUs in the group of Nitrosomonas ureae, and one OTU in the N. marina and N. aestuarii lineages, indicating the dominance of these species in the activated sludge samples. This almost perfectly matches the above results of this study using amoA gene as the biomarker. A minor exception is that Nitrospira 16S rRNA gene was not found by pyrosequencing, probably due to the low abundance of these AOB species in activated sludge and that the sequencing depth (>20,000 reads per sample) of the pyrosequencing applied in this study was not of sufficient depth to represent the complexity of the rare AOB sequences. Although we identified 495 sequence reads to the betaproteobacterial AOB, no reads can be assigned to the gammaproteobacterial AOB 16S rRNA genes, indicating the dominance of betaproteobacterial AOB in samples of this study.
Neighbor-joining phylogenetic tree based on AOB 16S rRNA gene sequences. The evolutionary distances were computed using the Jukes–Cantor method and are in the units of the number of base substitutions per site. Bootstrap values are indicated on branch nodes. Sequences obtained in this study are shown with “OTU-” in the names. Other sequences were obtained from GenBank
Abundance of AOB in activated sludge
The abundances of AOB in activated sludge were quantified using qPCR and pyrosequencing. However, the two data sets show a certain degree of discordance that might be due to the following reasons: (1) primer bias in qPCR amplification may introduce many uncertainties, especially when the target gene copy numbers are at low levels (Herrmann et al. 2008; Smith et al. 2006); (2) the 454 pyrosequencing conducted in this study may not be at a sufficient read depth to reflect the AOB complexity since AOB sequences was not detected in one of these samples; (3) the qPCR normalization step made use of total activated sludge DNA, which contains nucleic acids from eukaryotes, bacteria, archaea, and viruses to which any variations in the relative distribution of these components between samples would distort the qPCR normalization process; and (4) 16S rRNA gene copy numbers in prokaryotic microorganisms (Klappenbach et al. 2001) are different and the amoA gene copy numbers in different AOB are also different (Norton et al. 2002).
Diversity of AOA amoA gene in different activated sludge samples
Although many reports described the diversity and abundance of AOAs in the natural environments, only limited information is available describing AOA diversity in the activated sludge (Limpiyakorn et al. 2010; Park et al. 2006; Wells et al. 2009; Zhang et al. 2009). One possible reason for this situation could be due to the difficulty in PCR amplification of AOA amoA from DNA of sludge samples. In this study, only five out of eight samples were successfully amplified, and only three samples were eventually successfully cloned and sequenced.
Previous studies revealed that most of AOA should be assigned to the clusters of CGI.1a (the marine and sediment lineage) and CGI.1b (the soil lineage) in the phylum Crenarchaeota (Hatzenpichler et al. 2008). In this study, it was found that most of the AOA in the activated sludge samples have a closer distance to those AOA found in soil and thus belonged to the soil lineage. This result was in agreement with the previous studies on AOA communities in activated sludge (Limpiyakorn et al. 2010; Park et al. 2006), except for a study on the activated sludge in bioreactors treating saline sewage (Jin, et al. 2010) which had dominant AOA communities belonging to the marine and sediment lineage. AOA AmoA protein sequences were classified into nine OTUs using 3% distance cut-off. Figure S4 shows the relative abundance and distribution of AOA AmoA protein OTUs in different activated sludge samples, indicating lower diversity compared with AOA amoA gene and that most of the AOA AmoA protein OTUs were shared among the three samples. The neighbor-joining phylogenetic tree based on AOA AmoA protein sequences in Fig. S5 demonstrates the same pattern as shown by the AOA amoA gene tree (Fig. 6), that is, most of the OTUs in this study were grouped together and had a far distance from those sequences of marine and sediment. Interestingly, there are two OTUs (OTU-6 and OTU-7 in Fig. S5) closely related to the marine and sediment AOA lineage. The two OTUs were only detected in the sludge SG-SG-UP, a sample from Singapore, implying that the occurrence of these OTUs might be due to the geographic location of Singapore, which is near the ocean. The other two samples were taken from cities far from the ocean and thus their AOA populations mainly come from soil.
Neighbor-joining phylogenetic tree based on AOA amoA gene sequences. The evolutionary distances were computed using the Jukes–Cantor method and are in the units of the number of base substitutions per site. Bootstrap values are indicated on branch nodes. Sequences recovered in this study are shown with “OTU-” in the names. Other sequences were obtained from GenBank. The words in square brackets indicate the source the sequences originated from
References
Altschul S, Gish W, Miller W, Myers E, Lipman D (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Avrahami S, Liesack W, Conrad R (2003) Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ Microbiol 5(8):691–705
Claesson M, O'Sullivan O, Wang Q, Nikkila J, Marchesi J, Smidt H, De Vos W, Ross R, O'Toole P (2009) Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PloS One 4(8):e6669
DeSantis T, Hugenholtz P, Larsen N, Rojas M, Brodie E, Keller K, Huber T, Dalevi D, Hu P, Andersen G (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA 102(41):14683–14688
Good I (1953) The population frequencies of species and the estimation of population parameters. Biometrika 40(3–4):237–264
Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, Daims H, Wagner M (2008) A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA 105(6):2134–2139
Herrmann M, Saunders AM, Schramm A (2008) Archaea dominate the ammonia-oxidizing community in the rhizosphere of the freshwater macrophyte Littorella uniflora. Appl Environ Microbiol 74(10):3279–3283
Huson D, Auch A, Qi J, Schuster S (2007) MEGAN analysis of metagenomic data. Genome Res 17(3):377–386
Jin T, Zhang T, Yan Q (2010) Characterization and quantification of ammonia-oxidizing archaea (AOA) and bacteria (AOB) in a nitrogen-removing reactor using T-RFLP and qPCR. Appl Microbiol Biotechnol 87(3):1167–1176
Klappenbach J, Saxman P, Cole J, Schmidt T (2001) rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res 29(1):181–184
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4):299–306
Limpiyakorn T, Kurisu F, Yagi O (2006) Quantification of ammonia-oxidizing bacteria populations in full-scale sewage activated sludge systems and assessment of system variables affecting their performance. Water Sci Technol 54(1):91–99
Limpiyakorn T, Sonthiphand P, Rongsayamanont C, Polprasert C (2010) Abundance of amoA genes of ammonia-oxidizing archaea and bacteria in activated sludge of full-scale wastewater treatment plants. Bioresour Technol 102(4):3694–3701
Nicol G, Schleper C (2006) Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14(5):207–212
Norton J, Alzerreca J, Suwa Y, Klotz M (2002) Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch Microbiol 177(2):139–149
Park H, Noguera D (2004) Evaluating the effect of dissolved oxygen on ammonia-oxidizing bacterial communities in activated sludge. Water Res 38(14–15):3275–3286
Park H, Wells G, Bae H, Criddle C, Francis C (2006) Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl Environ Microbiol 72(8):5643–5647
Purkhold U, Pommerening-Roser A, Juretschko S, Schmid MC, Koops HP, Wagner M (2000) Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl Environ Microbiol 66(12):5368–5382
Robertson L, Kuenen J (1990) Combined heterotrophic nitrification and aerobic denitrification in Thiosphaera pantotropha and other bacteria. Antonie Van Leeuwenhoek 57(3):139–152
Rotthauwe JH, Witzel KP, Liesack W (1997) The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63(12):4704–4712
Schramm A, de Beer D, van den Heuvel J, Ottengraf S, Amann R (1999) Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol 65(8):3690–3696
Siripong S, Rittmann B (2007) Diversity study of nitrifying bacteria in full-scale municipal wastewater treatment plants. Water Res 41(5):1110–1120
Smith C, Nedwell D, Dong L, Osborn A (2006) Evaluation of quantitative polymerase chain reaction based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ Microbiol 8(5):804–815
Strous M, Fuerst J, Kramer E, Logemann S, Muyzer G, Van De Pas-Schoonen K, Webb R, Kuenen J, Jetten M (1999) Missing lithotroph identified as new planctomycete. Nature 400(6743):446–449
Tokutomi T, Shibayama C, Soda S, Ike M (2010) A novel control method for nitritation: the domination of ammonia oxidizing bacteria by high concentrations of inorganic carbon in an airlift-fluidized bed reactor. Water Res 44(4):4195–4203
Wells G, Park H, Yeung C, Eggleston B, Francis C, Criddle C (2009) Ammonia-oxidizing communities in a highly aerated full-scale activated sludge bioreactor: betaproteobacterial dynamics and low relative abundance of Crenarchaea. Environ Microbiol 11(9):2310–2328
Zhang T, Jin T, Yan QM, Shao MF, Wells G, Criddle CS, Fang HHP (2009) Occurrence of ammonia-oxidizing archaea (AOA) in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J Appl Microbiol 107:970–977
Acknowledgments
The authors wish to thank the Hong Kong General Research Fund (HKU7197/08E) for the financial support of this study. Lin Ye wishes to thank HKU for the postgraduate studentship. We would also like to thank W. Chan and C. K. Wong for technical help in pyrosequencing.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 2.28 mb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Zhang, T., Ye, L., Tong, A.H.Y. et al. Ammonia-oxidizing archaea and ammonia-oxidizing bacteria in six full-scale wastewater treatment bioreactors. Appl Microbiol Biotechnol 91, 1215–1225 (2011). https://doi.org/10.1007/s00253-011-3408-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3408-y