Abstract
Ideal immunogenicity in antigens is a prerequisite to eliciting a sufficiently strong immune and memory response via either DNA or protein vaccines. To improve immunogenicity, efforts have focused on high-level expression of target proteins and on maintaining their natural conformations. In the present work, two trimer motifs (MTQ and MTI) were designed and introduced into a plasmid vector with the tissue plasminogen activator signal peptide (tPA-SP). Next, we examined the efficacy and the efficiency of the two motifs as well as the introduction of tPA-SP and its mutant forms, 22P/A and 22P/G, in facilitating the secretory expression of trimeric proteins in mammalian cells. We found that both trimer motifs could produce the target protein in a trimeric form at a high level. Introduction of tPA-SP 22P/A markedly increased the secretory expression level. The combination of the trimer motif, MTQ, and the signal peptide, 22P/A, may serve as a universal mammalian vector for producing trimeric proteins in vaccine development.
Similar content being viewed by others
Introduction
To elicit a strong immune and memory response via DNA vaccines or DNA priming, ideal immunogenicity in antigens is a prerequisite (Kutzler and Weiner 2008). To improve immunogenicity, efforts have focused on a high-level expression of target proteins in vivo (Garmory et al. 2003) and on maintaining their natural conformations (Forsell et al. 2009; Wei et al. 2008).
Some viral immunogens, such as the HIV-1 envelope glycoprotein (Kwong et al. 2000), influenza virus hemagglutinin (Sammalkorpi and Lazaridis 2007), measles virus fusion protein (Navaratnarajah et al. 2009), and mumps virus fusion protein (Liu et al. 2006), naturally exist in a trimeric form that is necessary for proper function and immunogenicity. HIV-1 vaccine studies have shown that the oligomerised envelope antigens can elicit a stronger immune response than in their monomeric form (Beddows et al. 2005; Derby et al. 2006; Kim et al. 2005; Yang et al. 2001). Hence, mimicking the natural trimeric structure of an antigen is a primary concern for researchers to maintain the immunogenicity.
Proteins that naturally form a trimer have their own oligomerization domain within their polypeptide sequence. To maintain and stabilize the natural structure of such proteins in mammalian expression systems, researchers use a variety of methods. Such methods include the introduction of disulfide bonds to link the target protein with its natural oligomerization domain (Schulke et al. 2002), fusion expression of the target protein with its oligomerization domain (Yang et al. 2000b), and the introduction of a heterologous trimer motif into the target antigen (Bhardwaj et al. 2008; Hernandez Alvarez et al. 2008; Papanikolopoulou et al. 2004; Yang et al. 2000a). The last method has been demonstrated to be the most efficient and is widely utilized. The GCN4 leucine zipper (LZ), derived from yeast transcription factor GCN4 (Eckert et al. 1998; Harbury et al. 1993), is a widely used, trimeric coiled-coil model (Cornelissen et al. 2010; Yan et al. 2009; Yang et al. 2000b). Another commonly used trimer motif is FT, the foldon domain of the bacteriophage T4 fibritin (Bhardwaj et al. 2008; Papanikolopoulou et al. 2004; Yang et al. 2002). However, the trimeric proteins were observed in a non-preponderant proportion compared with their monomeric and dimeric forms (Yang et al. 2000a,b, 2002).
The expression of target proteins in vivo at high levels and in a secretory form will also ensure that ideal immunogenicity is maintained. A 23-bp signal sequence for the tissue plasminogen activator (tPA-SP) has been used as a heterologous leader sequence to drive a target protein into the cellular secretion pathway (Costa et al. 2006; Golden et al. 2008; Jalah et al. 2007; Kaur et al. 2009; Luo et al. 2008; Seo et al. 2009; Yang et al. 2001). Using sequence analysis, we found that the amino acid at position 22 of tPA-SP, position −1 of its cleavage site, is proline (P). However, it has been found that small polar residues with short, neutral side chains, such as alanine (A), glycine (G), serine (S), or threonine (T), are preferred at positions −1 and −3 proximal to the cleavage site in signal peptides (von Heijne 1983). Therefore, it is worth finding out whether substitution of 22P in tPA-SP with the amino acids mentioned above can improve the potency of tPA-SP in leading the secretory expression of proteins.
In the present study, a mammalian plasmid vector containing a new trimer motif and tPA-SP with 22A was designed and constructed to improve the secretory expression of trimeric proteins. We found that substitution of P at position 22 with a significantly improved the secretion of proteins. This motif can generate a high proportion of foreign proteins in their trimeric form.
Materials and methods
Design of the vectors
Two strategies were used in the vector construction. One was the introduction of either the tPA-SP sequence or one of its mutant forms, 22P/A and 22P/G, into the pcDNA3.1 plasmid (Invitrogen, USA) as a universal leading sequence to optimize the expression and secretion of heterologous proteins in mammalian cells. The two tPA-SP mutants, 22P/A and 22P/G, had an amino acid substitution at position 22P in tPA-SP with A or G, respectively. The other strategy was the introduction of a trimer motif, either MTQ (GGSGGIKEEIAKIKEEQAKIKEKIAEIEKRIAEIEKRIAGGCC) or MTI (GGSGGIKEEIAKIKEEIAKIKEKIAEIEKRIAEIEKRIAGGCC), which was designed, based on the coiled-coil model (Mason and Arndt 2004), to express the trimeric form of target proteins. A target gene was inserted between the C-terminus of the tPA-SP and the N-terminus of the trimer motif (Fig. 1).
Sequence prediction of the signal peptides and motifs
The likely cleavage efficiency of the signal peptides was predicted using the online prediction methods SignalP-HMM and SignalP-NN with the eukaryotic model (http://www.cbs.dtu.dk/services/SignalP/; Bendtsen et al. 2004; Nielsen et al. 1997). The secondary structure of the two motifs was predicted using the online program PSIPRED v2.6 (http://bioinf4.cs.ucl.ac.uk:3000/psipred/; Jones 1999; McGuffin et al. 2000). The probability for a coiled-coil structure was also predicted using the online program MARCOIL1.0 (http://www.isrec.isb-sib.ch/webmarcoil/webmarcoilC1.html; Delorenzi and Speed 2002).
Gene amplification and construction of the recombinant plasmids
The tPA-SP (GenBank accession no. E02360), MTQ, and MTI genes were synthesized by the Sangon Company (Shanghai, China) and then inserted into pcDNA3.1, as described above. The plasmids containing either 22P/A or 22P/G were constructed via site-directed mutagenesis.
An HIV-1 gag p24 gene was used as an example of a non-secretory protein, while the hemagglutinin glycoprotein subunit 1 (HA1) from the influenza virus H3N2 (ADCC VR-1679) was used as an example of a trimeric secretory protein. The HIV-1 CNHLJM06054 (GenBank accession no. EU131839) p24 gene was derived as described (Zhou et al. 2008). Viral RNA from the influenza virus was extracted using TRIzol Reagent (Invitrogen, USA), according to the manufacturer’s instruction. The HA1 complementary DNAs either containing (HA1.wt) or lacking the original signal sequence were amplified using RT-PCR. A gp120 gene (GenBank accession no. EU131805) (Zhang et al. 2007), which was codon-optimized according to the mammalian bias and lacked its original signal sequence, was synthesized by Sangon (Shanghai, China). One of the target genes was inserted into a plasmid either with or without a motif and either containing or lacking a signal peptide. All of the constructs are shown in Fig. 1.
Transient transfection
HEK 293T cells, which were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, USA) supplemented with 10% fetal calf serum, were transiently transfected with one of the plasmids listed in Fig. 1. All of the transfections were carried out using the Lipofectamine 2000 reagent (Invitrogen, USA) according to the instruction manual. Briefly, 5 × 105/well of 293T cells were plated in six-well plates and transfected with 4 μg of a plasmid in 2 ml of culture medium. Seventy-two hours later, the cell lysates and culture supernatants were collected. The total protein in each sample was quantitated using the BCA Protein Assay Kit (Pierce, USA). The expression levels of the target proteins were examined via Western blot and quantitative ELISA.
Western blot
The proteins expressed in the 293T cell lysates and supernatants were detected either by denaturing sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) or non-reducing PAGE and Western blot.
In the denaturing SDS-PAGE, samples containing equal quantities of total protein were treated in a sample buffer containing 1% β-mercaptoethanol (β-ME) and loaded on SDS-PAGE gels at equal volume. We also analyze the target proteins’oligomerization pattern mediated by either the trimer motif MTQ or MTI. For this analysis, the culture supernatants containing equal quantities of total protein were treated with non-reducing sample buffer (without β-ME) and loaded on the SDS-PAGE gels. After electrophoresis, the proteins were transferred onto PVDF membranes (Millipore, USA). The P24 proteins were detected using the monoclonal anti-P24 antibody VAK4 (Hattori et al. 1987) at a 1:500 dilution, followed by a horseradish peroxidise (HRP)-conjugated goat anti-mouse antibody (Zhongshan GoldenBridge Biotechnology, China) at a 1:2,000 dilution. The HA1 proteins were detected using a HA-Tag monoclonal antibody (Cell Signaling Technology, USA) at a 1:1,000 dilution, followed by an HRP-conjugated goat anti-rabbit antibody (Zhongshan GoldenBridge Biotechnology, China) at a 1:2,000 dilution. The gp120 proteins were detected using a polyclonal anti-gp120 rabbit serum (Advanced Biotechnologies Inc., USA) at a 1:1,000 dilution, followed by an HRP-conjugated goat anti-rabbit antibody (Zhongshan GoldenBridge Biotechnology, China) at a 1:2,000 dilution. The membranes were developed using the Enhanced Chemiluminescent Development System (Boster Company, Wuhan, China). The chemiluminescence signal was measured, and the intensity of each protein band from the Western blot result was scanned using an LAS4000 image analyzer (Fujifilm, Japan). The expression level of each target protein was shown as the relative expression to either P24T.tPA or HA1T.wt.
Quantitative p24 ELISA
The P24 expression levels in transfected 293T cell lysates and supernatants were determined by quantitative enzyme-linked immunosorbent assay (ELISA) using the HIV-1 p24 antigen ELISA kit (Zeptometrix, Buffalo, NY, USA) according to the instruction manual. Two independent transfections were performed. P24 protein in the supernatant and cell lysate from each transfection was measured twice.
Statistical analysis
A one-way ANOVA was performed for the statistical analysis. If statistical differences were observed in ANOVA, then a Student–Newman–Keuls post hoc test (SigmaStat 3.5 program) was employed to determine statistically significant differences between the two groups. P < 0.05 was considered statistically significant.
Results
tPA-SP with 22A rather than 22G significantly increased the secretion of heterologous proteins over the original tPA-SP
In most cases, the signal sequence is cleaved during or shortly after the polypeptide is transferred into the ER (Pfeffer and Rothman 1987). Improved cleavage of the leading sequence in a signal peptide may increase the secretion of the target protein (Chapman et al. 1991; Wang et al. 2006). Cleavage efficiency prediction using Model SignalP-HMM implied that the cleavage probabilities of the SP for tPA, 22P/A, and 22P/G were 0.686, 0.925, and 0.892, respectively (Fig. 2). Model SignalP-NN (data not shown) predicted a similar tendency, which suggested that substitution at this amino acid could generate a stronger cleavage tendency.
In the present study, we examined the efficacy of the tPA-SP 22P/A substitution in leading the expression and secretion of target proteins. We chose HIV-1 P24 and influenza virus hemagglutinin HA1 as models, as the former is non-secretory while the latter is a secretory protein. The expression of either P24 or HA1 in the cell lysates (CL) and supernatants (SN) from the transfected 293T cells was detected. Densitometry analysis was performed based on the intensity of the Western blot bands. The intensity of the P24T.tPA or HA1T.wt band was defined as 100%, and the expressions of the target proteins from the remaining constructs were shown as relative expression.
We observed the P24 protein in the cell lysate from each of the P24-expression vectors (Fig. 3a). Furthermore, there was no detectable P24 observed in the P24T-transfected supernatant. However, when the original tPA SP, the 22P/A SP, or the 22P/G SP led expression, P24 was detectable in the supernatant. This indicated that tPA-SP and its mutants directed the P24 into the secretion pathway. Compared with P24T.tPA, P24T.22P/A increased P24 levels by 65% (P < 0.001) in the supernatant and 62% (P < 0.001) in the cell lysate, while P24T.22P/G increased P24 levels only by 18% (P = 0.116) in the supernatant and 27% (P = 0.004) in the cell lysate.
The relative expression levels of the P24 and HA1 proteins that were directed by the various SPs. Expression analyses of the P24 (a) and HA1 (b) proteins from transiently transfected 293T cell lysates (CL) and supernatants (SN) using denaturing SDS-PAGE and Western blot are shown. The GAPDH was used as a reference protein and the levels of P24 or HA1 in CL were normalized to GAPDH. The Western blot results and the histograms were from one representative and three to four independent transfections, respectively. The data are presented as the mean ± standard error
Moreover, we observed HA1 expression that was directed by the HA1 wild-type signal sequence (HA1T.wt) (Fig. 3b). The SP from tPA, 22P/A, and 22P/G significantly increased HA1 expression (P < 0.05). Compared with HA1T.wt, HA1T.22P/A increased HA1 levels by 115% (P = 0.03) in the supernatant and 135% (P < 0.001) in the cell lysate; HA1T.22P/G increased HA1 levels by 90% (P = 0.005) in the supernatant and 81% (P < 0.001) in the cell lysate. The modification 22P/A in tPA-SP demonstrated higher expression levels than its parental sequence (P = 0.002 for the supernatant and P < 0.001 for the cell lysate), while the modification of 22P/G did not significantly increased HA1 expression in the supernatant compared with tPA-SP (P = 0.057)
Meanwhile, a quantitative ELISA was performed to determine the P24 levels in the supernatants and cell lysates (Table 1). The P24 levels in the two-milliliter transfected supernatant from P24T.tPA, P24T.22P/A, and P24T.22P/G were 6.67, 12.99, and 8.20 ng, respectively. Compared with the SP from tPA, the SP from 22P/A significantly increased the secretion of P24 (P < 0.001). The modification 22P/A resulted in the highest total level (15.86 ng) and highest secretion proportion (81.6%). P24T again failed in yielding a detectable level of P24 to the supernatant.
Introduction of the trimer motif MTQ generated a higher proportion of trimeric HA1 proteins than MTI
We designed two trimer motifs, MTQ and MTI, in accordance with features of the coiled-coil structure (Fig. 4). The core sequences for MTQ and MTI contained an obligatory helix with the highest confidence (Conf = 9), based on a prediction using the program PSIPRED v2.6. Similarly, prediction results from MARCOIL1.0 demonstrated that the core helix, E10 to K37, had a propensity of over 95% to form a coiled-coil structure, indicating that the four heptad repeats within MTQ and MTI tend to form an oligomerized structure (Woolfson 2005).
Secondary structure prediction of the trimer motifs. a The secondary structure predicted by the program PSIPRED v2.6. Conf Confidence of prediction (0 = low, 9 = high); Pred predicted secondary structures (H helix, C coil); AA, target sequence. b The secondary structure predicted by the program MARCOIL1.0
In the present work, we used the influenza virus HA1 as the target protein to examine the abilities of the two motifs to produce trimeric, heterologous proteins in the context of tPA-SP, as it naturally expresses as monomeric rather than a trimer. The HA1 secretion in the supernatants from the transiently transfected 293T cells was analysed using non-reducing PAGE followed by Western blot.
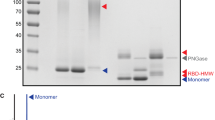
We found that when MTQ was fused to the C-terminus of HA1, the proportion of trimeric HA1 that was secreted into the supernatant from HA1T.wt, HA1T.tPA, HA1T.22P/A, and HA1T.22P/G was 72%, 74%, 75%, and 76%, respectively, compared with total HA1 (Fig. 5a, b). It suggested that more than 70% of the secretory HA1 protein was assembled into a trimeric form, aided by MTQ. The proportion of monomeric and dimeric HA1 that was expressed from these constructs ranged from 5% to 7% and 19% to 21%, respectively. In contrast, the constructs without the trimer motif resulted in no expression of trimeric HA1 (Fig. 5c).
The secretory expression of the HA1 proteins in the transiently transfected 293T cells. The expressions directed by MTQ (a, HA1T), MTI (b, HA1T.I), or no motif (c, HA1M) were examined using non-reducing PAGE and Western blot (upper panels) and the proportion of monomeric, dimeric and oligomeric HA1 proteins (lower panels) are shown. One representative result from three independent experiments is shown
On the other hand, although the introduction of the MTI motif aided in HA1 protein trimer assembly, the proportion of trimeric HA1 was much lower than that from the MTQ-containing construct (75% from HA1T.22P/A versus 56% from HA1T.I.22P/A) (Fig. 5b).
The vector bearing the MTQ motif and the tPA-SP of 22P/A yielded high-efficiency secretion of trimeric HIV-1 gp120
Introducing both tPA-SP 22P/A and the trimer motif MTQ yielded improved secretory expression and a high proportion of trimeric HA1. We still wondered whether these strategies could be applied to the expression of high-molecular-weight proteins. Thus, the ability of the expression system to produce trimeric HIV-1 gp120 glycoprotein was examined.
We first optimized the HIV-1 06044 gp120 codon according to the mammalian bias and subsequently constructed the recombinant plasmids gp120T.22P/A and gp120M.22P/A to express the monomeric and trimeric gp120 proteins, respectively. It was found that plasmid gp120M.22P/A produced only monomeric gp120 (Fig. 6a). In contrast, the gp120 protein from gp120T.22P/A was detected in monomeric, dimeric, and trimeric forms in the following proportions: 7%, 18%, and 75%, respectively (Fig. 6a, b). The proportion of gp120 trimer was similar to that of the trimeric HA1 protein from the HA1T-expression plasmids (Fig. 5). This suggested that the plasmid bearing the signal peptide 22P/A and the trimer motif MTQ could produce a high proportion of a trimeric high-molecular-weight protein.
The secretory expression of the gp120 protein in the transiently transfected 293T cells. The expression of gp120 with (gp120T) or without (gp120M) the trimer motif MTQ were examined using non-reducing PAGE and Western blot (a) and the proportion of monomeric, dimeric and oligomeric gp120 (b) are shown. One representative result from three independent experiments is shown
Discussion
The primary aim of this work was to improve the secretory expression level of trimeric proteins in transiently transfected mammalian cells. We used two strategies: the introduction of a stronger signal sequence and the utilization of a potent trimer motif.
The SP derived from tissue plasminogen activator was chosen in this work because of its reputation as one of the strongest leader sequences in secretory expression studies for many proteins and in various research fields (Chapman et al. 1991; Li et al. 1999; Qiu et al. 2000; Wang et al. 2006). It has been found that certain proteins, such as HIV-1 envelope glycoprotein, are secreted at low levels in protein expression systems in vitro or in DNA vaccine research. Furthermore, it was noted that such proteins have a marked increase in protein production after the substitution of their native SPs with tPA-SP (Chapman et al. 1991; Wang et al. 2006). In addition, the introduction of tPA-SP into certain proteins that lack SPs can naturally produce proteins in their secretory form and significantly improve the immunogenicity of protein-encoding DNA vaccines (Li et al. 1999; Midha and Bhatnagar 2009; Qiu et al. 2000).
We found in the present study that the wild-type HA1 signal peptide demonstrated a much weaker capacity than tPA-SP in leading secretory expression of HA1. Moreover, it is known that proper and efficient cleavage of signal peptides is the key element for further synthesis and folding of a nascent polypeptide into functional protein via the secretion pathway (Andrews et al. 1992; Coleman et al. 1985). The “−1/−3 rule” for signal peptides requires that certain preferential amino acid residues are at positions −1 to −3 proximal to the SP cleavage site (von Heijne 1983). We found that substitution of P with A at the −1 position in tPA-SP greatly increased the secretory expression of the target proteins that we examined, including P24 and HA1, although P24 is a naturally non-secretory protein. However, a G at the −1 position did not significantly increase the level of secretion. In addition to above results, cleavage efficiency prediction using both Model SignalP-HMM and Model SignalP-NN also suggested that the amino acid substitution 22P/A could generate the strongest cleavage tendency among all the SPs examined. These findings demonstrated that an A rather than a G at position 22 is the better design for tPA-SP. Further investigations as to the influence of amino acids, apart from the two investigated herein, on the cleavage efficiency is likely a meaningful step toward obtaining the best SP. To our knowledge, this is the first comparison study that examined the effect of amino acid preference within the tPA-SP cleavage site on protein expression and secretion.
The expression of viral proteins in mammalian cells has been demonstrated at extremely low levels, and the nanogram per milliliter level was reported previously (Alekseeva et al. 2009; Araujo et al. 2009; Haas et al. 1996). In HIV-1 protein expression, researchers have transiently transfected plasmids coding HIV-1 Gag protein, which disassembles into P24, P17, P7, and P6 protein for HIV-1 virion formation, and the P24 protein in the HIV-1 virion was found in the range of 0–3 ng/ml in transiently transfected mammalian cells (Deml et al. 2001; Qiu et al. 1999; zur Megede et al. 2000). This production level is consistent with our result using the P24T construct. However, such a level of secretory expressions is insufficient to produce enough protein to perform biological analysis and vaccine research, especially in large animal models. With respect to improving the efficacy of a signal peptide, great future efforts are still required.
For satisfactory immunogenicity and vaccine efficacy, both efficient expression of an antigen and maintenance of its natural conformation are necessary. However, producing stable trimeric proteins in vitro and in vivo is not always successful, although various approaches have been attempted (Beddows et al. 2007; Bhardwaj et al. 2008; Du et al. 2008; Harbury et al. 1993; Hernandez Alvarez et al. 2008; Papanikolopoulou et al. 2004; Schulke et al. 2002; Yang et al. 2000a,b; 2002). The key event for an ideal motif is the formation of a coiled-coil structure, which is known as the structural basis of an oligomeric conformation (Gonzalez et al. 1996; Harbury et al. 1993). Coiled coils are characterised by heptad repeats, (abcdefg) n , in which the buried a and d positions form the interface between multiple α-helices (Akey et al. 2001; Knappenberger et al. 2002). The GCN4 leucine zipper is a well-studied model system for three-strand coiled coils (Akey et al. 2001; Gonzalez et al. 1996; Harbury et al. 1993; Knappenberger et al. 2002) and has been used in leading the oligomerization of heterologous proteins (Cornelissen et al. 2010; Yan et al. 2009; Yang et al. 2000b). However, low trimer proportion (Yang et al. 2002) and the thermolability of trimeric proteins were reported in the previous research (Cornelissen et al. 2010; Yang et al. 2002). Based on these studies, we designed the two trimer motifs MTQ and MTI. The only difference between the two motifs was the amino acid residue at the a position in the second heptad repeat, where the buried a was glutamine (Q) for MTQ and isoleucine (I) for MTI (Fig. 4). It was shown by prediction using the programs PSIPRED and MARCOIL1.0 that the four heptad repeats in the core domain of either of the two motifs have a high tendency to form a coiled coil. This demonstrated, theoretically, the tendency of the motifs to generate a high proportion of oligomeric target proteins. However, we found from our expression tests that a higher proportion of trimers among the total HA1 could be driven by MTQ, while the motif MTI yielded more dimerization and less trimerization than MTQ.
Polar or charged residues have been observed at the a and d positions, although typically hydrophobic residues are located here (Akey et al. 2001; Mason and Arndt 2004). Furthermore, certain polar residues, such as Q, tyrosine (Y), S, and T, in the interface affect not only local packing but also overall coiled coil geometry (Akey et al. 2001; Wagschal et al. 1999). In the initial design of the motifs for the present study, we attempted to maintain the hydrophobic core of the coiled coils with Ile at positions a and d to form the oligomerized structure. In addition, we introduced a polar residue, Q, at the a position in the second heptad repeat to see if it may enhance the tendency toward trimerization. We have successfully achieved this goal. We do not yet know the reason why Q can improve the proportion of trimer expression in the MTQ motif, but it may be important for oligomerization specificity and chain orientation in a trimeric coiled-coil formation (Gratkowski et al. 2001; Wagschal et al. 1999). Furthermore, it might be sensible to examine the remaining polar residues to define which is best.
The efficacy of the trimer motif MTQ has been verified by the results of our transient protein expressions and the structure predictions. However, we still do not know whether the trimeric form reflects the native conformation of the target proteins. This question would be answered by the results of a crystallographic analysis. Additionally, whether such a trimeric protein can maintain its natural activity and succeed in satisfactory immunogenicity must also be confirmed in the future.
Taken together, we have demonstrated that the tPA signal peptide with 22P/A can successfully improve the secretory expression of foreign proteins and that the trimer motif MTQ may drive a high proportion of trimeric proteins. We also demonstrated that a polar residue at the a position of the second heptad repeat within the coiled coil improved the trimerization of target proteins. The combination of tPA-SP 22P/A with the motif MTQ could be considered a “double-optimized” design for secretory expression of trimeric proteins and serve as a universal mammalian vector.
References
Akey DL, Malashkevich VN, Kim PS (2001) Buried polar residues in coiled-coil interfaces. Biochemistry 40(21):6352–6360
Alekseeva E, Sominskaya I, Skrastina D, Egorova I, Starodubova E, Kushners E, Mihailova M, Petrakova N, Bruvere R, Kozlovskaya T, Isaguliants M, Pumpens P (2009) Enhancement of the expression of HCV core gene does not enhance core-specific immune response in DNA immunization: advantages of the heterologous DNA prime, protein boost immunization regimen. Genet Vaccines Ther 7:7
Andrews DW, Young JC, Mirels LF, Czarnota GJ (1992) The role of the N region in signal sequence and signal-anchor function. J Biol Chem 267(11):7761–7769
Araujo NM, Vianna CO, Moraes MT, Gomes SA (2009) Expression of Hepatitis B virus surface antigen (HBsAg) from genotypes A, D and F and influence of amino acid variations related or not to genotypes on HBsAg detection. Braz J Infect Dis 13(4):266–271
Beddows S, Franti M, Dey AK, Kirschner M, Iyer SP, Fisch DC, Ketas T, Yuste E, Desrosiers RC, Klasse PJ, Maddon PJ, Olson WC, Moore JP (2007) A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 360(2):329–340
Beddows S, Schulke N, Kirschner M, Barnes K, Franti M, Michael E, Ketas T, Sanders RW, Maddon PJ, Olson WC, Moore JP (2005) Evaluating the immunogenicity of a disulfide-stabilized, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 79(14):8812–8827
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340(4):783–795
Bhardwaj A, Walker-Kopp N, Wilkens S, Cingolani G (2008) Foldon-guided self-assembly of ultra-stable protein fibers. Protein Sci 17(9):1475–1485
Chapman BS, Thayer RM, Vincent KA, Haigwood NL (1991) Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res 19(14):3979–3986
Coleman J, Inukai M, Inouye M (1985) Dual functions of the signal peptide in protein transfer across the membrane. Cell 43(1):351–360
Cornelissen LA, de Vries RP, de Boer-Luijtze EA, Rigter A, Rottier PJ, de Haan CA (2010) A single immunization with soluble recombinant trimeric hemagglutinin protects chickens against highly pathogenic avian influenza virus H5N1. PLoS ONE 5(5):e10645
Costa SM, Paes MV, Barreto DF, Pinhao AT, Barth OM, Queiroz JL, Armoa GR, Freire MS, Alves AM (2006) Protection against dengue type 2 virus induced in mice immunized with a DNA plasmid encoding the non-structural 1 (NS1) gene fused to the tissue plasminogen activator signal sequence. Vaccine 24(2):195–205
Delorenzi M, Speed T (2002) An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18(4):617–625
Deml L, Bojak A, Steck S, Graf M, Wild J, Schirmbeck R, Wolf H, Wagner R (2001) Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J Virol 75(22):10991–11001
Derby NR, Kraft Z, Kan E, Crooks ET, Barnett SW, Srivastava IK, Binley JM, Stamatatos L (2006) Antibody responses elicited in macaques immunized with human immunodeficiency virus type 1 (HIV-1) SF162-derived gp140 envelope immunogens: comparison with those elicited during homologous simian/human immunodeficiency virus SHIVSF162P4 and heterologous HIV-1 infection. J Virol 80(17):8745–8762
Du C, Wang M, Liu J, Pan M, Cai Y, Yao J (2008) Improvement of thermostability of recombinant collagen-like protein by incorporating a foldon sequence. Appl Microbiol Biotechnol 79(2):195–202
Eckert DM, Malashkevich VN, Kim PS (1998) Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues. J Mol Biol 284(4):859–865
Forsell MN, Schief WR, Wyatt RT (2009) Immunogenicity of HIV-1 envelope glycoprotein oligomers. Curr Opin HIV AIDS 4(5):380–387
Garmory HS, Brown KA, Titball RW (2003) DNA vaccines: improving expression of antigens. Genet Vaccines Ther 1(1):2
Golden JW, Josleyn MD, Hooper JW (2008) Targeting the vaccinia virus L1 protein to the cell surface enhances production of neutralizing antibodies. Vaccine 26(27–28):3507–3515
Gonzalez L Jr, Woolfson DN, Alber T (1996) Buried polar residues and structural specificity in the GCN4 leucine zipper. Nat Struct Biol 3(12):1011–1018
Gratkowski H, Lear JD, DeGrado WF (2001) Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci USA 98(3):880–885
Haas J, Park EC, Seed B (1996) Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol 6(3):315–324
Harbury PB, Zhang T, Kim PS, Alber T (1993) A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 262(5138):1401–1407
Hattori T, Sagawa K, Matsushita S, Koito A, Suto H, Matsuoka M, Yokoyama M, Takatsuki K (1987) Characterization of three monoclonal antibodies (VAK3-5) that identify p24, core protein of human immunodeficiency virus, and its precursors. Jpn J Cancer Res 78(3):235–241
Hernandez Alvarez B, Hartmann MD, Albrecht R, Lupas AN, Zeth K, Linke D (2008) A new expression system for protein crystallization using trimeric coiled-coil adaptors. Protein Eng Des Sel 21(1):11–18
Jalah R, Rosati M, Kulkarni V, Patel V, Bergamaschi C, Valentin A, Zhang GM, Sidhu MK, Eldridge JH, Weiner DB, Pavlakis GN, Felber BK (2007) Efficient systemic expression of bioactive IL-15 in mice upon delivery of optimized DNA expression plasmids. DNA Cell Biol 26(12):827–840
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292(2):195–202
Kaur M, Rai A, Bhatnagar R (2009) Rabies DNA vaccine: no impact of MHC class I and class II targeting sequences on immune response and protection against lethal challenge. Vaccine 27(15):2128–2137
Kim M, Qiao ZS, Montefiori DC, Haynes BF, Reinherz EL, Liao HX (2005) Comparison of HIV Type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res Hum Retroviruses 21(1):58–67
Knappenberger JA, Smith JE, Thorpe SH, Zitzewitz JA, Matthews CR (2002) A buried polar residue in the hydrophobic interface of the coiled-coil peptide, GCN4-p1, plays a thermodynamic, not a kinetic role in folding. J Mol Biol 321(1):1–6
Kutzler MA, Weiner DB (2008) DNA vaccines: ready for prime time? Nat Rev Genet 9(10):776–788
Kwong PD, Wyatt R, Sattentau QJ, Sodroski J, Hendrickson WA (2000) Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J Virol 74(4):1961–1972
Li Z, Howard A, Kelley C, Delogu G, Collins F, Morris S (1999) Immunogenicity of DNA vaccines expressing tuberculosis proteins fused to tissue plasminogen activator signal sequences. Infect Immun 67(9):4780–4786
Liu Y, Xu Y, Lou Z, Zhu J, Hu X, Gao GF, Qiu B, Rao Z, Tien P (2006) Structural characterization of mumps virus fusion protein core. Biochem Biophys Res Commun 348(3):916–922
Luo M, Tao P, Li J, Zhou S, Guo D, Pan Z (2008) Immunization with plasmid DNA encoding influenza A virus nucleoprotein fused to a tissue plasminogen activator signal sequence elicits strong immune responses and protection against H5N1 challenge in mice. J Virol Methods 154(1–2):121–127
Mason JM, Arndt KM (2004) Coiled coil domains: stability, specificity, and biological implications. Chembiochem 5(2):170–176
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405
Midha S, Bhatnagar R (2009) Anthrax protective antigen administered by DNA vaccination to distinct subcellular locations potentiates humoral and cellular immune responses. Eur J Immunol 39(1):159–177
Navaratnarajah CK, Leonard VH, Cattaneo R (2009) Measles virus glycoprotein complex assembly, receptor attachment, and cell entry. Curr Top Microbiol Immunol 329:59–76
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8(5–6):581–599
Papanikolopoulou K, Forge V, Goeltz P, Mitraki A (2004) Formation of highly stable chimeric trimers by fusion of an adenovirus fiber shaft fragment with the foldon domain of bacteriophage t4 fibritin. J Biol Chem 279(10):8991–8998
Pfeffer SR, Rothman JE (1987) Biosynthetic protein transport and sorting by the endoplasmic reticulum and Golgi. Annu Rev Biochem 56:829–852
Qiu JT, Liu B, Tian C, Pavlakis GN, Yu XF (2000) Enhancement of primary and secondary cellular immune responses against human immunodeficiency virus type 1 gag by using DNA expression vectors that target Gag antigen to the secretory pathway. J Virol 74(13):5997–6005
Qiu JT, Song R, Dettenhofer M, Tian C, August T, Felber BK, Pavlakis GN, Yu XF (1999) Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J Virol 73(11):9145–9152
Sammalkorpi M, Lazaridis T (2007) Configuration of influenza hemagglutinin fusion peptide monomers and oligomers in membranes. Biochim Biophys Acta 1768(1):30–38
Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC (2002) Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol 76(15):7760–7776
Seo SH, Jin HT, Park SH, Youn JI, Sung YC (2009) Optimal induction of HPV DNA vaccine-induced CD8+ T cell responses and therapeutic antitumor effect by antigen engineering and electroporation. Vaccine 27(42):5906–5912
von Heijne G (1983) Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem 133(1):17–21
Wagschal K, Tripet B, Lavigne P, Mant C, Hodges RS (1999) The role of position a in determining the stability and oligomerization state of alpha-helical coiled coils: 20 amino acid stability coefficients in the hydrophobic core of proteins. Protein Sci 8(11):2312–2329
Wang S, Farfan-Arribas DJ, Shen S, Chou TH, Hirsch A, He F, Lu S (2006) Relative contributions of codon usage, promoter efficiency and leader sequence to the antigen expression and immunogenicity of HIV-1 Env DNA vaccine. Vaccine 24(21):4531–4540
Wei CJ, Xu L, Kong WP, Shi W, Canis K, Stevens J, Yang ZY, Dell A, Haslam SM, Wilson IA, Nabel GJ (2008) Comparative efficacy of neutralizing antibodies elicited by recombinant hemagglutinin proteins from avian H5N1 influenza virus. J Virol 82(13):6200–6208
Woolfson DN (2005) The design of coiled-coil structures and assemblies. Adv Protein Chem 70:79–112
Yan Z, Holmes KV, Hodges RS (2009) Expression and characterization of recombinant S2 subunit of SARS-coronavirus S fusion protein. Adv Exp Med Biol 611:153–154
Yang X, Farzan M, Wyatt R, Sodroski J (2000a) Characterization of stable, soluble trimers containing complete ectodomains of human immunodeficiency virus type 1 envelope glycoproteins. J Virol 74(12):5716–5725
Yang X, Florin L, Farzan M, Kolchinsky P, Kwong PD, Sodroski J, Wyatt R (2000b) Modifications that stabilize human immunodeficiency virus envelope glycoprotein trimers in solution. J Virol 74(10):4746–4754
Yang X, Lee J, Mahony EM, Kwong PD, Wyatt R, Sodroski J (2002) Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J Virol 76(9):4634–4642
Yang X, Wyatt R, Sodroski J (2001) Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J Virol 75(3):1165–1171
Zhang W, Zhou HZ, Wang KL, Cheng DC, Zhang FM, Ling H (2007) Analyzing of the genetic characteristics of HIV-1 CHLJBF06044 envelope. J Harbin Med Univ 41(6):537–540
Zhou HZ, Li Y, Zhou H, Liu YC, Du HT, Wang KL, Wang FX, Ling H (2008) A new CRF01_AE/B recombinant structure of HIV type 1 found in Heilongjiang province, China. AIDS 22(13):1690–1693
zur Megede J, Chen MC, Doe B, Schaefer M, Greer CE, Selby M, Otten GR, Barnett SW (2000) Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J Virol 74(6):2628–2635
Acknowledgments
The authors are grateful to Dr. Bin Wang at the China Agricultural University for his careful reading of the manuscript and his comments as well as Dr. Hattori T at the Tohoku University of Japan for kindly providing the monoclonal antibody VAK4. This work was supported by the National Natural Science Foundation of China (30771910), the National Grand Program on Key Infectious Disease, Ministry of Health of China (2008ZX10001-012), the Natural Science Foundation of Heilongjiang Province (ZJY0601-01), and the Heilongjiang Innovation Program in Graduate Education (YJSCX2009-222HLJ).
Author information
Authors and Affiliations
Corresponding author
Additional information
J.-Y. Wang and W.-T. Song contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, JY., Song, WT., Li, Y. et al. Improved expression of secretory and trimeric proteins in mammalian cells via the introduction of a new trimer motif and a mutant of the tPA signal sequence. Appl Microbiol Biotechnol 91, 731–740 (2011). https://doi.org/10.1007/s00253-011-3297-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3297-0