Abstract
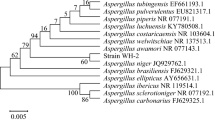
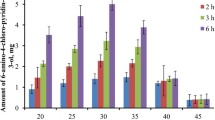
A novel bacterial strain HS0904 was isolated from a soil sample using 3,5-bis(trifluoromethyl) acetophenone as the sole carbon source. This bacterial isolate can asymmetrically reduce 3,5-bis(trifluoromethyl) acetophenone to (1R)-[3,5-bis(trifluoromethyl)phenyl] ethanol with high enantiometric excess (ee) value. Based on its morphological, physiological characteristics, Biolog, 16S rDNA sequence and phylogenetic analysis, strain HS0904 was identified as Leifsonia xyli HS0904. To our knowledge, this is the first reported case on the species L. xyli exhibited R-stereospecific carbonyl reductase and used for the preparation of chiral (1R)-[3,5-bis(trifluoromethyl)phenyl] ethanol. The optimization of parameters for microbial transformation of 3,5-bis(trifluoromethyl) acetophenone to (1R)-[3,5-bis(trifluoromethyl)phenyl] ethanol catalyzed by whole cells of L. xyli HS0904 was carried out by examining some key factors including buffer pH, reaction temperature, shaking speed, substrate concentration, and reaction time. The obtained optimized conditions for the bioreduction are as follows: buffer pH 8.0, 70 mM of 3,5-bis(trifluoromethyl) acetophenone, 100 g l−1 of glucose as co-substrate, 200 g l−1 of resting cells as biocatalyst, reaction for 30 h at 30 °C and 200 rpm. Under above conditions, 99.4% of product ee and best yield of 62% were obtained, respectively. The results indicated that isolate L. xyli HS0904 is a novel potential biocatalyst for the production of (1R)-[3,5-bis(trifluoromethyl)phenyl] ethanol.








Similar content being viewed by others
References
Bazhanov DP, Yatsevich KK, Bazhanova AA (2010) Phylogenetic identification of three strains of Rhizosphere bacteria based on the results of 16s rRNA gene analysis and genetic typing. Microbiology 79:374–384
Brands KMJ, Payack JF, Rosen JD, Nelson TD (2003) Efficient synthesis of NK1 receptor antagonist aprepitant using a crystallization-induced diastereoselective transformation. J Am Chem Soc 125:2129–2135
Gelo-Pujic M, Guyader FL, Schlama T (2006) Microbial and homogenous asymmetric catalysis in the reduction of 1-[3,5-bis(trifluoromethyl)phenyl] ethanone. Tetrahedron: Asymmetry 17:2000–2005
Genov DG, Ager DJ (2004) Asymmetric hydrogenation of ketones catalyzed by RuII–bicp complexes. Angew Chem Int Ed 43:2816–2819
Hansen KB, Chilenski JR, Desmond R, Devine PN (2003) Scalable, efficient process for the synthesis of (R)-3,5-bistrifluoromethylphenyl ethanol via catalytic asymmetric transfer hydrogenation and isolation as a DABCO inclusion complex. Tetrahedron: Asymmetry 14:3581–3587
Homann MJ, Vail RB, Previte E, Tamarez M, Morgan B, Dodds DR, Zaks A (2004) Rapid identification of enantioselective ketone reductions using targeted microbial libraries. Tetrahedron 60:789–797
Huisman GW, Liang J, Krebber A (2010) Practical chiral alcohol manufacture using ketoreductases. Curr Opin Chem Biol 14:122–129
Ju X, Yu HL, Pan J, Wei DZ, Xu JH (2010) Bioproduction of chiral mandelate by enantioselective deacylation of α-acetoxyphenylacetic acid using whole cells of newly isolated Pseudomonas sp. ECU1011. Appl Microbiol Biotechnol 86:83–91
Kaluzna IA, Rozzell JD, Kambourakis S (2005) Ketoreductases: stereoselective catalysts for the facile synthesis of chiral alcohols. Tetrahedron: Asymmetry 16:3682–3689
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformatics 9:299–306
Kurbanoglu EB, Zilbeyaz K, Taskin M, Kurbanoglu NI (2009) Total production of (R)-3,5-bistrifluoromethylphenyl ethanol by a symmetric reduction of 3,5-bis(trifluoromethyl)-acetophenone in the submerged culture of Penicillium expansum isolate. Tetrahedron: Asymmetry 20:2759–2763
Kurbanoglu EB, Zilbeyaz K, Ozdal M, Taskin M, Kurbanoglu NI (2010) Asymmetric reduction of substituted acetophenones using once immobilized Rhodotorula glutinis cells. Bioresour Technol 101:3825–3829
Li W, Sun XF, Zhou L, Hou GH, Yu SC, Zhang XM (2009) Highly efficient and highly enantioselective asymmetric hydrogenation of ketones with tunesphos/1,2-diamine ruthenium(II) complexes. J Org Chem 74:1397–1399
Matsuda T, Yamanaka R, Nakamura K (2009) Recent progress in biocatalysis for asymmetric oxidation and reduction. Tetrahedron: Asymmetry 20:513–557
Pollard D, Truppo M, Pollard J, Chen CY, Moore J (2006) Effective synthesis of (S)-3,5-bistrifluoromethylphenyl ethanol by asymmetric enzymatic reduction. Tetrahedron: Asymmetry 17:554–559
Susumu N, Tomoya O, Junsaku K (2008) Population pharmacokinetics of aprepitant and dexamethasone in the prevention of chemotherapy-induced nausea and vomiting. Cancer Chemother Pharmacol 63:75–83
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Vankawala PJ, Kolla N, Elati CR (2007) Efficient synthesis of (1R)-[3,5-bis(trifluoromethyl)phenyl] ethanol, a key intermediate for aprepitant, an NK-1 receptor antagonist. Synthetic Commun 37:3439–3446
Wang P, Sun LM, He JY (2009) Medium optimization for enhanced production of carbonyl reductase by Candida tropicalis 104 by response surface methodology. Chin J Biotechnol 25:863–868
Wilson K (1997) Preparation of genomic DNA from bacteria. In: Ausubel FM, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) Current protocols in molecular biology. Wiley, New York, pp 241–245
Xie Y, Xu JH, Xu Y (2010) Isolation of a Bacillus strain producing ketone reductase with high substrate tolerance. Bioresour Technol 101:1054–1059
Zerillo MM, Van Sluys MA, Camargo LE, Monteiro-Vitorello CB (2008) Characterization of new IS elements and studies of their dispersion in two subspecies of Leifsonia xyli. BMC Microbiol 8:127
Acknowledgments
This work was financially supported by grants from National Natural Science Foundation (21076193), People's Republic of China, and the Foundation of Zhejiang Key Developing Discipline of Pharmacy (20100609), Zhejiang Province, People's Republic of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, P., Cai, JB., Ouyang, Q. et al. Asymmetric biocatalytic reduction of 3,5-bis(trifluoromethyl) acetophenone to (1R)-[3,5-bis(trifluoromethyl)phenyl] ethanol using whole cells of newly isolated Leifsonia xyli HS0904. Appl Microbiol Biotechnol 90, 1897–1904 (2011). https://doi.org/10.1007/s00253-011-3233-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3233-3