Abstract
Synthetic biology is the attempt to apply the concepts of engineering to biological systems with the aim to create organisms with new emergent properties. These organisms might have desirable novel biosynthetic capabilities, act as biosensors or help us to understand the intricacies of living systems. This approach has the potential to assist the discovery and production of pharmaceutical compounds at various stages. New sources of bioactive compounds can be created in the form of genetically encoded small molecule libraries. The recombination of individual parts has been employed to design proteins that act as biosensors, which could be used to identify and quantify molecules of interest. New biosynthetic pathways may be designed by stitching together enzymes with desired activities, and genetic code expansion can be used to introduce new functionalities into peptides and proteins to increase their chemical scope and biological stability. This review aims to give an insight into recently developed individual components and modules that might serve as parts in a synthetic biology approach to pharmaceutical biotechnology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
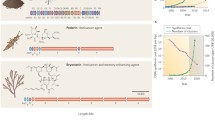
Nature provides a reservoir of active compounds that help us to fight and cure disease (Fig. 1). We have learned to exploit this reservoir by identifying and isolating the active compounds, by developing strategies for their chemical synthesis and by improving their biological activity. However, this process is laborious and very expensive. The large-scale isolation of drugs is often hindered by the low amount of material produced by the natural source, and the complexity of the molecules makes their total chemical synthesis costly to unprofitable. Transferring the ability to biosynthesize these compounds from the source organisms to genetically friendlier hosts would greatly facilitate their production. Genome sequencing efforts now provide the genetic information of entire metabolic pathways necessary to recombinantly produce pharmaceutically relevant molecules. However, biosynthetic pathways are often highly intertwined, regulated by extensive feedback mechanisms and require the correct organization of the individual entities in space and time. The simple transfer of genetic information from one organism to another is therefore rarely sufficient to confer the biosynthetic capability. Thus, we need to understand the properties of individual components as well as the properties of entire systems and eventually learn to recombine artificial biosynthetic networks in new hosts in a predictable, efficient manner. The design of life with novel properties from individual modules in a way resembling the creation of new machines in modern engineering is the ultimate aim of the emergent discipline of synthetic biology.
Molecular structure of members of important natural drug families. a Vancomycin, a cyclo-peptide antibiotic. b Cyclosporin, a family member of the cyclic peptides and an important immunosuppressant. c Inhibitor of HIV budding selected from an in vitro cyclic peptide library (Tavassoli et al. 2008). d Artemisinin, a terpenoid with anti-malarial activity. e Epothilone D, member of the polyketide family of natural products with cytostatic activity
In its most stringent definition, synthetic biology is the application of the principles of engineering to the construction of life with desired properties in a rational and systematic way (Serrano 2007). Synthetic biology therefore uses well-characterized, modular parts that can be put together to create new functionality in a directed, predictable manner (Kiel et al. 2010; Ninfa 2010; Topp and Gallivan 2010). Several recent advances in DNA technology encourage the followers of this new discipline: First, inexpensive DNA synthesis enables the facile production of codon-usage optimized genes and other genetic elements, such as promoters and regulatory sequences. This technology has recently been improved towards the synthesis of an entire genome, which was subsequently used to substitute the genome of a living cell (Gibson et al. 2009; Gibson et al. 2008a, b; Lartigue et al. 2007, 2009). Second, rapid, next-generation DNA sequencing technologies provide vast amounts of genetic information. Entire genome sequences of many organisms are now publicly available and can be mined electronically for relevant genes (Benson et al. 2009). Third, well-characterized genetic modules are being developed, which can be combined rationally to create a new function (Lu et al. 2009). The Registry of Standard Biological Parts (http://parts.mit.edu) is a continuously growing collection of standardized parts (“BioBricks”) that are being used to rationally create organisms with desired properties (Voigt 2006). Directed evolution has become a widespread strategy to adapt existing parts to novel functions. For example, the directed evolution of tRNAs and amino acyl-tRNA synthetases (aaRS) to decode novel, unnatural amino acids has been used to expand the genetic code of several organisms (Chin et al. 2003; Liu et al. 2007; Wang et al. 2001) and allows genetically programming the incorporation of unprecedented functionality into proteins in vivo (Xie and Schultz 2006).
With increasing experience in the reprogramming of microorganisms, entirely new types of therapy might become feasible in the future. Bacteria might no longer be limited to the mere production of drugs but will also be able to deliver them to the site of action. In this review, we want to give an overview of recently developed individual components and of the attempts to recombine them for the production of modules of biotechnological and pharmaceutical interest. We are trying to give an impression of how these individual modules might eventually be used as parts in a synthetic biology approach to pharmaceutical biotechnology.
Creating in vivo compound libraries
Combinatorial small molecule libraries are an efficient way to search for new lead structures. Establishing these libraries synthetically, however, is costly and many initial hits unfortunately fail in subsequent rounds of screening or in clinical trials.
Producing compound libraries directly in vivo would have several advantages. First, maintaining and amplifying such a library is simple and can be done by cultivating cells. Second, the screening process is facilitated because the genetic information remains directly coupled to the compound. Hence, the selected compound can be identified by analysing the genetic information of the corresponding cell. Third, an intracellular genetic selection can directly assay for effects on enzymatic activity, bypassing inherent limitations of in vitro assays. Fourth, the selection is performed in the context of a living cell requiring an enhanced level of selectivity of the drug for its target. Finally, potential problems with solubility and uptake of the compound are circumvented.
Cyclic peptide libraries
Many important natural compounds are modified cyclic peptides, for example, antibiotics like vancomycin (Fig. 1a), the immunosuppressant drug cyclosporin (Fig. 1b) and fungal toxins phalloidin or actinomycin D (Katsara et al. 2006). Cyclization renders peptides resistant to cellular degradation and restricts conformational freedom, potentially improving binding affinity and specificity. Using a split intein approach, the Benkovic lab has developed a method to produce libraries of cyclic peptides in Escherichia coli (Scott et al. 1999). A number of promising candidates have since been identified in libraries generated this way (Horswill et al. 2004; Tavassoli and Benkovic 2005; Tavassoli et al. 2008). Natural cyclic peptides contain many modified, non-coding amino acids. This increases the diversity of these compounds and is crucial for their biological activity. As detailed below, the artificial expansion of the genetic code allows the co-translational incorporation of unnatural amino acids (Wang et al. 2001; Xie and Schultz 2006). Combining the Benkovic approach to the production of cyclic peptides with the incorporation of unnatural amino acids would vastly increase the diversity of cyclic peptides that can be produced in vivo.
Polyketide synthases—natural synthetic biology
Another important class of natural products are the polyketides. Polyketides are synthesized by large multi-enzyme complexes, the polyketide synthases. These “assembly lines” are built from modular components that catalyze the formation of the carbon chain of the final product in a stepwise manner (Fig. 2). The modular composition of polyketide synthases makes them an ideal playground for synthetic biologists. Their individual modules can be split and recombined to form active hybrid enzymes (Watanabe et al. 2003b). The combinatorial recombination of individual modules has been achieved, and the new enzymes were shown to successfully catalyze the formation of polyketides in E. coli (Menzella et al. 2005). This approach can potentially be used to produce libraries of polyketides with novel biological activities in vivo. Presently, simple recombination of different modules often produces inactive synthases because the transfer of intermediates between modules might be blocked or the connectivity between modules disturbed. Growing information on the structure of individual modules and their connectivity (Alekseyev et al. 2007; Keatinge-Clay and Stroud 2006; Tang et al. 2006) together with increasing experimental experience will help to develop predictive algorithms to rationally design synthases for “unnatural” polyketides in a combinatorial biosynthetical approach (Khosla et al. 2009). If a system to produce large libraries of hybrid enzymes is connected to a selectable output (Yin et al. 2007), active clones can be identified, even if their frequency in the library is very low (Menzella and Reeves 2007). A combination of computer-assisted prediction, combinatorial library design guided by structural information and selection might eventually develop into the mainstream of drug discovery.
Microbial polyketide synthases are modular assembly lines that fit polyketides together from monomeric building blocks. In the first step of the reaction, the starter module is acylated with the first unit (1). This unit is subsequently transferred to the first of a row of extender modules that catalyze the elongation of the polyketide. All extender modules contain at least a ketosynthase (KS), an acyl transferase (AT) and an acyl carrier protein (ACP) domain. In each step, the KS receives the acyl group unit from the ACP of the preceding module and the AT adds an appropriate extender unit from its CoA ester to the prosthetic group of the ACP (2). The KS then catalyzes a decarboxylative Claisen condensation between the acylated KS and the extender unit to give a β-keto-acylated acyl carrier protein (3). Other modules may be included to further modify the growing polyketide chain. Upon completion of the elongation process, the polyketide chain is cleaved from the polyketide synthase by the thioesterase (TE) of the termination module (4)
De novo creation of metabolic pathways
The de novo design of biosynthetic pathways to expand the biosynthetic potential of microorganisms is an intriguing route towards compounds for which natural pathways have not been elucidated or which are not of natural origin (Prather and Martin 2008). A “retro-biosynthetic” approach, analogous to retrosynthesis in organic chemistry, uses enzymes with evolved substrate specificity to create artificial biosynthetic pathways de novo. Many natural enzymes show high selectivity for their substrates, which is the result of divergent evolution from promiscuous precursor proteins (O'Brien and Herschlag 1999). However, there are examples of natural enzymes with broad substrate specificity. Starting from such a promiscuous enzyme, the sesquiterpene synthase γ-humulene synthase, Keasling and co-workers were able to recapitulate this evolutionary process and create seven specific and active enzymes that use different reaction pathways and produce different products (Yoshikuni et al. 2006). These enzymes could be used in the future to design biosynthetic pathways for unnatural terpenoids.
Similarly, prenylation is an important modification occurring on natural products, like naphterpin, conferring anti-cancer, anti-viral or anti-microbial activity to the molecules (Botta et al. 2005). Prenyltransferases, the enzymes responsible for the derivatization, have been identified and characterized (Kuzuyama et al. 2005). These enzymes have a broad substrate spectrum and might form a starting point for the evolution of enzymes for regio-specific prenylation of aromatic small molecules. Engineered enzymes might eventually allow us to create enzymatic pathways de novo and to produce small molecule libraries of differently modified compounds of the same scaffold structure (Dietrich et al. 2009; Yoshikuni et al. 2006).
Expanding the chemistry of life
The cellular environment imposes a constraint to the chemical scope of reactions that can be used to create in vivo compound libraries. Many reactions familiar to synthetic organic chemists require elevated temperature or are incompatible with aqueous environments. This limitation may be overcome in the future by the computer-assisted, directed evolution of enzymes (Kaplan and DeGrado 2004) that catalyze unnatural chemical reactions. Using several enzymes as scaffolds, Baker and co-workers (Rothlisberger et al. 2008) employed computer-aided design to engineer enzymes able to catalyze a Kemp elimination reaction for which no natural enzyme is yet known. It is thus conceivable that the limits to the chemistries available in vivo will be shifted in the future, thereby expanding the chemical scope of in vivo compound libraries.
The diversity in the chemical nature of proteins and peptides has been tremendously increased by the invention of strategies to introduce unnatural amino acids in vivo. Peter Schultz and his co-workers have artificially expanded the genetic code of several organisms (Chin et al. 2003; Liu et al. 2007; Wang et al. 2001) and succeeded in incorporating a wide range of unnatural amino acids (Xie and Schultz 2006). The basic principle of genetic code expansion is the expression of an additional pair of tRNA and its cognate aaRS in the host system (Fig. 3). Both, tRNA and aaRS, must not be substrates for the host aaRSs and tRNAs to ensure correct translation of host proteins. Under these prerequisites, the tRNA can be mutated to decode amber stop codons, for example, and the aaRS can be evolved to specifically recognize unnatural amino acids. This system allows the incorporation of additional, unnatural amino acids in response to amber stop codons. In the past decade, the list of unnatural amino acids that can be incorporated in this way has grown continuously (Neumann et al. 2008; Nguyen et al. 2009b; Xie and Schultz 2006). Recent developments of amber and quadruplet suppressor ribosomes now allow the efficient incorporation of multiple different unnatural amino acids into the same protein (Neumann et al. 2010; Wang et al. 2007). These unnatural amino acids have side chains with unusual chemical properties and reactivities and could be used to increase the chemical diversity of peptides and proteins.
The principle of genetic code expansion. The basic principle of genetic code expansion is the heterologous expression of an additional tRNA/aminoacyl-tRNA synthetase (aaRS) pair in the host cell. This pair is orthogonal to the host’s translational machinery, meaning that the tRNA is not a substrate for endogenous synthetases and that the aaRS does not charge host tRNAs. The orthogonal tRNA is mutated to decode amber stop codons and the orthogonal aaRS is evolved to specifically recognize an unnatural amino acid (UAA) supplied with the growth medium. Cells with such an expanded genetic code are able to produce proteins with the UAA incorporated at a position encoded by an amber codon in the mRNA
Drug screening
The quality of an assay system has an enormous impact on the outcome of a screen. In order to fully profit from the advantages of in vivo compound libraries, it is necessary to develop intracellular assays, like a colour signal or a genetically selectable phenotype, to detect or select positive clones. The continuously increasing number of biosensors could serve as such detector modules.
The Benkovic group has developed a method for identifying small molecule inhibitors of protein–protein interactions by combining a reverse two-hybrid system with the split intein-mediated formation of cyclic peptide libraries (Horswill et al. 2004). Using this system, cyclic peptides were identified that modulate the interaction between the two subunits of ribonucleotide reductase (Horswill et al. 2004), HIV-Gag and TSG101 (Fig. 1c) (Tavassoli et al. 2008) and AICAR transformylase homodimerization (Tavassoli and Benkovic 2005). A similar system that combines in vivo cyclic peptide libraries with a fluorescent reporter assay was developed by the same group and used to identify antibacterial cyclic peptide inhibitors of ClpXP protease (Cheng et al. 2007).
Tsien and co-workers created the Ca2+-sensor “Cameleon” (Miyawaki et al. 1997) by fusing calmodulin to a calmodulin-binding peptide and flanking the construct with two fluorescent proteins with overlapping spectral properties. Ca2+-dependent binding of calmodulin to its own peptide results in a predictable conformational rearrangement that leads to a change in energy transfer between the two fluorescent proteins. Biosensors of this sort are being used extensively to monitor the intracellular concentration and dynamics of ions and small molecules and the activation state of signalling proteins (Balla 2009). Similar sensors could be designed for metabolites of interest and combined with systems to produce small molecule libraries in vivo in order to identify strains and conditions that allow production of pharmacologically relevant compounds.
Optimization of engineered metabolic pathways depends on our ability to measure the production of intermediates efficiently. Mass spectrometry can be used to quantify the relevant metabolites, but faster real-time assays allow the identification of optimal growth condition or the screening of mutant libraries in high-throughput formats. Mevalonate, for example, is an important precursor in the biosynthesis of isoprenoids. Keasling and co-workers (Pfleger et al. 2007) engineered an auxotrophic strain of E. coli into a mevalonate biosensor by expressing GFP. The strain reports on the mevalonate concentration in the growth medium through a change in growth rate. This biosensor strain can now be used to measure mevalonate in high-throughput formats.
In a more direct approach, reporter proteins could be created by redesigning the affinities of binding proteins, like maltose binding protein (MBP), for ligands of interest. MBP is a member of the periplasmic binding protein superfamily that mediates chemosensory processes in a wide range of bacteria. This protein family has evolved to bind a variety of ligands, such as amino acids, carbohydrates and ions, naturally and has served as starting point for the directed evolution of biosensors, receptors and enzymes (Dwyer and Hellinga 2004). Upon ligand binding, these proteins undergo a significant conformational change (induced fit), which has been exploited to monitor the event by positioning environmentally sensitive fluorophores within the binding site (endosteric placement), at the rim of the binding site (peristeric placement) or in a crevice that opens and closes with ligand binding (allosteric placement) (de Lorimier et al. 2002). Alternatively, GFP variants have been conjugated to the N- and C-termini of periplasmic-binding proteins in order to monitor ligand binding by fluorescence energy transfer (Fehr et al. 2002; Fehr et al. 2003; Schultz and Moini 2003). Computational design has been successfully employed to create biosensor proteins with novel affinities for small molecules, such as trinitrotoluene, L-lactate and serotonin (Looger et al. 2003). These engineered sensors were incorporated into synthetic bacterial signal transduction pathways to regulate gene expression in response to extracellular stimuli (Looger et al. 2003).
G-protein coupled receptors (GPCRs) are the target of the majority of pharmaceuticals. This membrane protein family is stimulated by extracellular ligands and initiates a signalling cascade on the cytosolic side of the membrane upon agonist binding. The study of GPCRs in their natural environment is usually complicated by the presence of multiple different receptors on a single cell combined with the tremendous efforts necessary for its genetic manipulation. Pausch and colleagues transplanted a mammalian GPCR into an engineered Saccharomyces cerevisiae strain and connected it to a downstream cascade controlling the expression of the his3 gene (Price et al. 1995). Activation of the receptor permitted the cells to grow in the absence of histidine. This approach has since been used extensively to study GPCR function and to identify ligands and antagonists (Brown et al. 2000; Minic et al. 2005; Sachpatzidis et al. 2003).
These examples demonstrate that it is possible to create modules for the detection of a molecule of interest by rational design. A synthetic biology approach to the production of pharmaceuticals should employ these sensors to control the expression level of individual parts or connect them to a selectable output. In a case study, Farmer and Liao coupled the E. coli Ntr regulon, which senses the intracellular metabolite acetylphosphate, to the expression of key enzymes in the pathway for the recombinant biosynthesis of lycopene (Farmer and Liao 2000). This approach significantly enhanced lycopene production and simultaneously reduced the toxic side effects caused by metabolic imbalance. Riboswitches, RNA elements with the ability to control the translation of a coupled open reading frame upon binding specifically to certain ligands, are another class of parts that can be used to control metabolism (Famulok et al. 2007). Even before their discovery in nature, artificial riboswitches had been designed to control the expression of reporter genes in response to the binding of small molecules, such as Hoechst dyes or tetracycline (Hanson et al. 2003; Suess et al. 2004; Suess et al. 2003; Werstuck and Green 1998). These elements could be used to couple gene expression on the translational level to the metabolic state of the cell. Similar strategies might form the basis of high-throughput screening systems to identify engineered strains that are able to biosynthesize the molecule of interest.
Biosynthesis of pharmaceuticals
Recombinant DNA technology has been used for more than three decades to engineer bacteria with the ability to produce pharmacologically important molecules like insulin (Goeddel et al. 1979). Progress has been made in recent years towards the heterologous production of important natural products, like terpenoids, polyketides, non-ribosomal peptides and alkaloids. Synthetic biology approaches are being employed to fine-tune the levels and activities of individual steps and components. Here, we present examples of recent successes of this strategy.
Terpenoids
Terpenoids are a large and diverse class of natural products with important applications in human health. Well-known examples are the anti-malarial drug artemisinin (Fig. 1d) (Tan et al. 1999), the anti-cancer agent paclitaxel (taxol) (Jennewein and Croteau 2001) and eleutherobin (Long et al. 1998). A major step forward towards the recombinant production of terpenoids in E. coli was the creation of a platform strain that heterologously expressed the mevalonate pathway of S. cerevisiae (Martin et al. 2003). The metabolic precursors produced by this strain can be further converted into relevant terpenoids by co-expression of the corresponding terpene synthases and modifying enzymes. Building on this work, the Keasling lab has engineered E. coli to produce artemisinic acid, which can be converted chemically into artemisinin (Roth and Acton 1989), with yields of more than 300 mg/l (Chang et al. 2007; Kizer et al. 2008; Newman et al. 2006). Similarly, the same group has recently reprogrammed the metabolism of S. cerevisiae to produce artemisinic acid with titres exceeding 100 mg/l (Ro et al. 2006).
Taxol (paclitaxel) is a well-established anti-neoplastic drug active against a variety of different cancers (Goldspiel 1997). Although the total synthesis of Taxol has been achieved (Kingston et al. 2002), the approach is too expensive for it being commercially practical (Borman 1994). The drug is produced by yew (Taxus) species in about 19 individual enzymatic steps from the isoprenoid precursor geranylgeranyl diphosphate (Jennewein and Croteau 2001; Walker and Croteau 2001). Several Taxol biosynthetic genes have been expressed functionally in S. cerevisiae and used to establish the five-step biosynthesis of an intermediate of Taxol (Dejong et al. 2006; Engels et al. 2008). Efforts are also being undertaken to establish Taxol biosynthesis in E. coli (Huang et al. 2001). However, there are still a number of problems to overcome, like the lack of a complete list of enzymes involved in the production of many important terpenoids, in order to achieve their heterologous total biosynthesis. Still, the achievements are promising and there is hope for cost-efficient production of some very important natural products in the near future.
Polyketides
Polyketides are a large class of important natural products, e.g. erythromycin, epothilone (Fig. 1e) and FK-506. They are produced by microorganisms, most prominently actinomycetes, soil-dwelling bacteria. A significant achievement in the recombinant production of polyketides in E. coli (that naturally does not produce any) was the engineering of the pathway that encodes the production of 6-deoxyerythronolide B (6dEB) (Pfeifer et al. 2001). This strain was created by optimizing precursor production, enzyme engineering and destroying catabolic pathways. Subsequent optimization of the strain has improved the level of 6dEB production to levels obtained from optimized S. coelicolor strains (Pfeifer et al. 2002). Building on this work, heterologous production of several other important polyketides or their precursors has been achieved recently, namely, the anti-cancer drugs epothilone C and D (Mutka et al. 2006) and ansamycin precursors (Rude and Khosla 2006; Watanabe et al. 2003a), aklanoic acid (precursor to several antitumor polyketides, e.g. doxorubicin and aclacinomycin A) (Lee et al. 2005) and aromatic bacterial polyketides (Zhang et al. 2008).
Metabolic fine-tuning
These promising examples illustrate the potential of a synthetic biology approach to the recombinant production of pharmaceutics. There are, however, significant challenges ahead. In many cases, the relevant precursor metabolites are inefficiently produced by the host or bottlenecks are formed during the synthesis of the product when an intermediate is unstable or an enzyme of low activity in the foreign environment. Synthetic biology has the aim to create autonomous circuits for the production of key metabolites allowing the modular combination of part reactions into new pathways. The carefully adjusted expression level of individual components of the pathway is important to optimize flux and to avoid the accumulation of toxic intermediates. Synthetic promoter libraries and a range of engineered constitutive promoters of varying strengths have been created (Alper et al. 2005; Hammer et al. 2006) and could be used to fine-tune the expression levels of all enzymes in a pathway in order to optimize metabolic flux.
The Church lab has recently published a novel technology for multiplex automated genome engineering (“MAGE”) to optimize metabolic pathways in E. coli. By feeding cells with synthetic oligonucleotides, which are probably used as Okazaki fragments during replication, the authors were able to mutate and fine-tune ribosomal binding sites genome-wide (Wang et al. 2009). They have used this method to increase the production of lycopene in an engineered E. coli strain by a factor of five within only 3 days of evolution. The same approach might allow optimizing the production of other metabolites if a simple screening assay is available. The use of an automated setup greatly accelerated and increased the throughput of this technology. The development of similar automated systems will have a major impact on the de novo creation of biosynthetic pathways and regulatory networks in the future.
An important reoccurring theme of cellular metabolism is compartmentalization and channelling of metabolites (Ovadi and Srere 2000). This avoids side reactions (especially hydrolysis) of activated intermediates and prevents toxic side effects. A strategy that mimics this organization of enzymes would facilitate the design of efficient metabolic pathways. A first step towards this aim has recently been taken: By fusing the components of the metabolic pathway for mevalonate to peptide motifs, Keasling and co-workers (Dueber et al. 2009) were able to recruit them into synthetic protein scaffolds and to optimize their stoichiometry within the synthetic complex. The activity of the pathway was thereby increased by a factor of 77. Compartmentalization (for example, in protein-based organelles that exist in many bacteria (Yeates et al. 2008)) can also help to establish new chemical reactions by shielding unstable intermediates from a hostile environment or by creating a special phase, e.g. with low or high pH or hydrophobicity.
Bacteria as drug delivery units
In recent years, we have witnessed an explosion of new technologies to manipulate cellular genetic information. Our engineering abilities allow us to design cells with artificial genomes (Gibson et al. 2009; Gibson et al. 2008a, b; Lartigue et al. 2007, 2009), promising the advent of customized drug-producing or therapeutic bacteria. It might become possible to equip “synthetic” organisms with just the genetic information necessary for function, while omitting any interfering and potentially dangerous genetic and metabolic material. These cells could be rationally equipped with modular genetic elements to create an organism with a desired phenotype. In a futuristic scenario, genetic devices might be combined to program organisms like robots. First attempts produced newly combined biosensor modules under newly linked genetic circuits (Kobayashi et al. 2004; Looger et al. 2003; Lu et al. 2009; You et al. 2004). For example, a chimeric sensor domain of a phytochrome from cyanobacteria fused to a signal transduction domain of E. coli was used to design an E. coli strain responding to light (Levskaya et al. 2005). Engineering new bacterial sensors to regulate genes in response to a new environmental context can be extended to create other sensors and also reprogram downstream signalling at wish. Promoters, for example, could be built to integrate (multiple) specific signals (Anderson et al. 2007).
This approach can be extended to design live bacteria as targeted delivery systems for live vaccination, as probiotics and anti-tumour agents (Garmory et al. 2003; Martin et al. 2003; Nguyen et al. 2010; Pawelek et al. 2003). Various bacteria invade tumours and have been engineered to destruct them by releasing a chemotherapeutic prodrug (Nemunaitis et al. 2003), via secretion of TNFα (Dang et al. 2001; Nuyts et al. 2001) and cytokines (Murray et al. 1996). However, a synthetic biological approach would allow designing a bacterium rationally with desired capabilities and safety requirements. The Voigt lab constructed an E. coli strain that senses a low-oxygen microenvironment such as found in tumour tissues. Hypoxia is the cue in this particular E. coli strain to upregulate a Yersinia pseudotuberculosis adhesin protein (invasin) sufficient for E. coli to invade mammalian cells. Because invasion is only efficient from a certain cell density, the Voigt lab further equipped this E. coli with a second sensor including another genetic circuit that senses cell density (quorum-sensing circuit from Vibrio fischeri) (Anderson et al. 2006). Thus, this E. coli strain should specifically invade tumour cells in a population density-dependent manner.
The type III secretion apparatus, usually used by certain bacteria to pump effector proteins into the eukaryotic host cytoplasm for manipulation, was exploited in a synthetic biology approach to pump spider silk proteins into the medium (Widmaier et al. 2009). Combined with the previous approaches, a (synthetic) bacterial strain equipped with rationally linked sensors and genetic circuits might be able to sense the environment of damaged tissues, specifically to adhere and release therapeutic proteins into the infected cell and thus could kill (or heal) damaged or infected tissues. Refining such approaches together with biological “safety tasks”, which can also be synthetic, would be a further step towards the use of safe therapeutic bacteria.
Optimizing drugs
Often the identification of a new drug candidate is only the beginning of a laborious and time-consuming effort to optimize and mature its pharmacokinetic properties.
Small therapeutic proteins, for example, have a short residence time in the bloodstream. This can be improved by modifying surface residues with polyethylene glycol (PEG). Random derivatization usually creates an ill-defined mixture of molecules with varying degrees of PEGylation. A strategy to introduce PEG residues in a defined way is therefore much preferable over random modification. This is being tried using incorporation of unnatural amino acids with an alkyne side chain (Deiters et al. 2004). Subsequent modification with azide group-containing PEG residues generates proteins with site-specific PEGylation.
Many proteins are stabilized by disulfide cross-bridges. In recombinant proteins, these disulfide bonds are usually difficult to reproduce because of their redox sensitivity and the lack of relevant chaperones. A general method for the genetically programmed installation of directed redox-insensitive cross-links has recently been devised (Neumann et al. 2010). The authors combined evolved orthogonal ribosomes with two pairs of mutually orthogonal tRNA/aaRS pairs to incorporate two different unnatural amino acids into the same protein. The side chains of these amino acids contained an azide and an alkyne functional group that formed a cross-link upon addition of a Cu(I) catalyst.
Post-translational modifications (PTMs) are often present on proteins and can tremendously change their properties. However, recombinant proteins from E. coli are usually unmodified. It is therefore often necessary to use eukaryotic expression systems to produce correctly processed proteins, but even then the pattern of modifications is usually different from the natural situation. Lysine acetylation has been recognized as a widespread PTM in prokaryotes and eukaryotes (Choudhary et al. 2009; Kim et al. 2006; Zhang et al. 2009). The effect of the modification on protein function is largely unexplored. A method to genetically encode this PTM on recombinant proteins in E. coli has recently been devised (Neumann et al. 2008; Neumann et al. 2009). Hence, therapeutic proteins harbouring this modification can now be produced recombinantly without any knowledge of the modifying enzymes involved. Similar methods exist to produce proteins containing lysine methylation or tyrosine sulphation (Liu and Schultz 2006; Nguyen et al. 2009a). Other PTMs that cannot be encoded genetically to date, like glycosylations or ubiquitination, can be mimicked by incorporating a chemical handle into the protein at the site of the modification followed by in vitro derivatization (Liu et al. 2003).
Conclusion and perspective
In this review, we did not intend to give a complete list of all the parts available to a synthetic biology approach to pharmaceutical biotechnology and we apologize to those whose work was left unmentioned (please see Kiel et al. (2010), Ninfa (2010) and Topp and Gallivan (2010) for more recent reviews of this topic.) Our aim was to give a flavour of the potential impact that synthetic biology might have on the pharmaceutical industry. Future work should use these parts to create systems for the simultaneous production, identification and optimization of new drugs. Such an approach might look as follows: A reporter is engineered to detect a protein–protein interaction or an enzymatic activity of interest. To this end, one could develop biosensors that sense a reaction product or genetic circuits that are activated once a certain interaction is prevented. Such a sophisticated system will reduce the number of false hits during the screening process. This reporter system is subsequently combined with various in vivo compound libraries to isolate clones with the desired activity. Newly identified lead structures can be further improved afterwards, for example, with the help of unnatural amino acids. Their biosynthesis could be optimized for instance by using MAGE or by compartmentalizing the enzymes involved. Any improvement to the system could be identified or selected quickly by the reporter. A lot of work need to be invested before such an approach will become the mainstream of drug development. An important next step towards this aim would be to form well-characterized modular parts from individual components that can be freely and quickly combined in the true sense of a synthetic biology approach to pharmaceutical biotechnology. The automation of such an approach combined with the development of computational design tools for synthetic biology (Marchisio and Stelling 2009) will be an important technological advance that will help in its realization.
References
Alekseyev VY, Liu CW, Cane DE, Puglisi JD, Khosla C (2007) Solution structure and proposed domain domain recognition interface of an acyl carrier protein domain from a modular polyketide synthase. Protein Sci 16:2093–2107
Alper H, Fischer C, Nevoigt E, Stephanopoulos G (2005) Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA 102:12678–12683
Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2006) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355:619–627
Anderson JC, Voigt CA, Arkin AP (2007) Environmental signal integration by a modular AND gate. Mol Syst Biol 3:133
Balla T (2009) Green light to illuminate signal transduction events. Trends Cell Biol 19:575–586
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2009) GenBank. Nucleic Acids Res 37:D26–D31
Borman S (1994) Total synthesis of anticancer agent taxol achieved by 2 different routes. Chem Eng News 72:32–34
Botta B, Vitali A, Menendez P, Misiti D, Delle Monache G (2005) Prenylated flavonoids: pharmacology and biotechnology. Curr Med Chem 12:717–739
Brown AJ, Dyos SL, Whiteway MS, White JH, Watson MA, Marzioch M, Clare JJ, Cousens DJ, Paddon C, Plumpton C, Romanos MA, Dowell SJ (2000) Functional coupling of mammalian receptors to the yeast mating pathway using novel yeast/mammalian G protein alpha-subunit chimeras. Yeast 16:11–22
Chang MC, Eachus RA, Trieu W, Ro DK, Keasling JD (2007) Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol 3:274–277
Cheng L, Naumann TA, Horswill AR, Hong SJ, Venters BJ, Tomsho JW, Benkovic SJ, Keiler KC (2007) Discovery of antibacterial cyclic peptides that inhibit the ClpXP protease. Protein Sci 16:1535–1542
Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG (2003) An expanded eukaryotic genetic code. Science 301:964–967
Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840
Dang LH, Bettegowda C, Huso DL, Kinzler KW, Vogelstein B (2001) Combination bacteriolytic therapy for the treatment of experimental tumors. Proc Natl Acad Sci USA 98:15155–15160
de Lorimier RM, Smith JJ, Dwyer MA, Looger LL, Sali KM, Paavola CD, Rizk SS, Sadigov S, Conrad DW, Loew L, Hellinga HW (2002) Construction of a fluorescent biosensor family. Protein Sci 11:2655–2675
Deiters A, Cropp TA, Summerer D, Mukherji M, Schultz PG (2004) Site-specific PEGylation of proteins containing unnatural amino acids. Bioorg Med Chem Lett 14:5743–5745
Dejong JM, Liu Y, Bollon AP, Long RM, Jennewein S, Williams D, Croteau RB (2006) Genetic engineering of taxol biosynthetic genes in Saccharomyces cerevisiae. Biotechnol Bioeng 93:212–224
Dietrich JA, Yoshikuni Y, Fisher KJ, Woolard FX, Ockey D, McPhee DJ, Renninger NS, Chang MC, Baker D, Keasling JD (2009) A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450(BM3). ACS Chem Biol 4:261–267
Dueber JE, Wu GC, Malmirchegini GR, Moon TS, Petzold CJ, Ullal AV, Prather KL, Keasling JD (2009) Synthetic protein scaffolds provide modular control over metabolic flux. Nat Biotechnol 27:753–759
Dwyer MA, Hellinga HW (2004) Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr Opin Struct Biol 14:495–504
Engels B, Dahm P, Jennewein S (2008) Metabolic engineering of taxadiene biosynthesis in yeast as a first step towards taxol (paclitaxel) production. Metab Eng 10:201–206
Famulok M, Hartig JS, Mayer G (2007) Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 107:3715–3743
Farmer WR, Liao JC (2000) Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol 18:533–537
Fehr M, Frommer WB, Lalonde S (2002) Visualization of maltose uptake in living yeast cells by fluorescent nanosensors. Proc Natl Acad Sci USA 99:9846–9851
Fehr M, Lalonde S, Lager I, Wolff MW, Frommer WB (2003) In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors. J Biol Chem 278:19127–19133
Garmory HS, Leary SE, Griffin KF, Williamson ED, Brown KA, Titball RW (2003) The use of live attenuated bacteria as a delivery system for heterologous antigens. J Drug Target 11:471–479
Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA 3rd (2008a) One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci USA 105:20404–20409
Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA 3rd, Smith HO (2008b) Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319:1215–1220
Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345
Goeddel DV, Kleid DG, Bolivar F, Heyneker HL, Yansura DG, Crea R, Hirose T, Kraszewski A, Itakura K, Riggs AD (1979) Expression in Escherichia coli of chemically synthesized genes for human insulin. Proc Natl Acad Sci USA 76:106–110
Goldspiel BR (1997) Clinical overview of the taxanes. Pharmacotherapy 17:110S–125S
Hammer K, Mijakovic I, Jensen PR (2006) Synthetic promoter libraries—tuning of gene expression. Trends Biotechnol 24:53–55
Hanson S, Berthelot K, Fink B, McCarthy JE, Suess B (2003) Tetracycline-aptamer-mediated translational regulation in yeast. Mol Microbiol 49:1627–1637
Horswill AR, Savinov SN, Benkovic SJ (2004) A systematic method for identifying small-molecule modulators of protein–protein interactions. Proc Natl Acad Sci USA 101:15591–15596
Huang Q, Roessner CA, Croteau R, Scott AI (2001) Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg Med Chem 9:2237–2242
Jennewein S, Croteau R (2001) Taxol: biosynthesis, molecular genetics, and biotechnological applications. Appl Microbiol Biotechnol 57:13–19
Kaplan J, DeGrado WF (2004) De novo design of catalytic proteins. Proc Natl Acad Sci USA 101:11566–11570
Katsara M, Tselios T, Deraos S, Deraos G, Matsoukas MT, Lazoura E, Matsoukas J, Apostolopoulos V (2006) Round and round we go: cyclic peptides in disease. Current Medicinal Chemistry 13:2221–2232
Keatinge-Clay AT, Stroud RM (2006) The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure 14:737–748
Khosla C, Kapur S, Cane DE (2009) Revisiting the modularity of modular polyketide synthases. Curr Opin Chem Biol 13:135–143
Kiel C, Yus E, Serrano L (2010) Engineering signal transduction pathways. Cell 140:33–47
Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23:607–618
Kingston DG, Jagtap PG, Yuan H, Samala L (2002) The chemistry of taxol and related taxoids. Fortschr Chem Org Naturst 84:53–225
Kizer L, Pitera DJ, Pfleger BF, Keasling JD (2008) Application of functional genomics to pathway optimization for increased isoprenoid production. Appl Environ Microbiol 74:3229–3241
Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ (2004) Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci USA 101:8414–8419
Kuzuyama T, Noel JP, Richard SB (2005) Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435:983–987
Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA 3rd, Smith HO, Venter JC (2007) Genome transplantation in bacteria: changing one species to another. Science 317:632–638
Lartigue C, Vashee S, Algire MA, Chuang RY, Benders GA, Ma L, Noskov VN, Denisova EA, Gibson DG, Assad-Garcia N, Alperovich N, Thomas DW, Merryman C, Hutchison CA 3rd, Smith HO, Venter JC, Glass JI (2009) Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325:1693–1696
Lee TS, Khosla C, Tang Y (2005) Engineered biosynthesis of aklanonic acid analogues. J Am Chem Soc 127:12254–12262
Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA (2005) Synthetic biology: engineering Escherichia coli to see light. Nature 438:441–442
Liu CC, Schultz PG (2006) Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol 24:1436–1440
Liu H, Wang L, Brock A, Wong CH, Schultz PG (2003) A method for the generation of glycoprotein mimetics. J Am Chem Soc 125:1702–1703
Liu W, Brock A, Chen S, Schultz PG (2007) Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods 4:239–244
Long BH, Carboni JM, Wasserman AJ, Cornell LA, Casazza AM, Jensen PR, Lindel T, Fenical W, Fairchild CR (1998) Eleutherobin, a novel cytotoxic agent that induces tubulin polymerization, is similar to paclitaxel (taxol). Cancer Res 58:1111–1115
Looger LL, Dwyer MA, Smith JJ, Hellinga HW (2003) Computational design of receptor and sensor proteins with novel functions. Nature 423:185–190
Lu TK, Khalil AS, Collins JJ (2009) Next-generation synthetic gene networks. Nat Biotechnol 27:1139–1150
Marchisio MA, Stelling J (2009) Computational design tools for synthetic biology. Curr Opin Biotechnol 20:479–485
Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802
Menzella HG, Reeves CD (2007) Combinatorial biosynthesis for drug development. Curr Opin Microbiol 10:238–245
Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV (2005) Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotechnol 23:1171–1176
Minic J, Persuy MA, Godel E, Aioun J, Connerton I, Salesse R, Pajot-Augy E (2005) Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening. FEBS J 272:524–537
Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388:882–887
Murray PJ, Aldovini A, Young RA (1996) Manipulation and potentiation of antimycobacterial immunity using recombinant bacille Calmette–Guerin strains that secrete cytokines. Proc Natl Acad Sci USA 93:934–939
Mutka SC, Carney JR, Liu Y, Kennedy J (2006) Heterologous production of epothilone C and D in Escherichia coli. Biochemistry 45:1321–1330
Nemunaitis J, Cunningham C, Senzer N, Kuhn J, Cramm J, Litz C, Cavagnolo R, Cahill A, Clairmont C, Sznol M (2003) Pilot trial of genetically modified, attenuated Salmonella expressing the E. coli cytosine deaminase gene in refractory cancer patients. Cancer Gene Ther 10:737–744
Neumann H, Peak-Chew SY, Chin JW (2008) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat Chem Biol 4:232–234
Neumann H, Hancock SM, Buning R, Routh A, Chapman L, Somers J, Owen-Hughes T, van Noort J, Rhodes D, Chin JW (2009) A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell 36:153–163
Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW (2010) Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464:441–444
Newman JD, Marshall J, Chang M, Nowroozi F, Paradise E, Pitera D, Newman KL, Keasling JD (2006) High-level production of amorpha-4, 11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol Bioeng 95:684–691
Nguyen DP, Garcia Alai MM, Kapadnis PB, Neumann H, Chin JW (2009a) Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones. J Am Chem Soc 131:14194–14195
Nguyen DP, Lusic H, Neumann H, Kapadnis PB, Deiters A, Chin JW (2009b) Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA synthetase/tRNA(CUA) pair and click chemistry. J Am Chem Soc 131:8720–8721
Nguyen VH, Kim HS, Ha JM, Hong Y, Choy HE, Min JJ (2010) Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res 70(1):18–23
Ninfa AJ (2010) Use of two-component signal transduction systems in the construction of synthetic genetic networks. Curr Opin Microbiol 13(2):240–245
Nuyts S, Van Mellaert L, Theys J, Landuyt W, Bosmans E, Anne J, Lambin P (2001) Radio-responsive recA promoter significantly increases TNFalpha production in recombinant clostridia after 2 Gy irradiation. Gene Ther 8:1197–1201
O'Brien PJ, Herschlag D (1999) Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol 6:R91–R105
Ovadi J, Srere PA (2000) Macromolecular compartmentation and channeling. Int Rev Cytol 192:255–280
Pawelek JM, Low KB, Bermudes D (2003) Bacteria as tumour-targeting vectors. Lancet Oncol 4:548–556
Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C (2001) Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790–1792
Pfeifer B, Hu Z, Licari P, Khosla C (2002) Process and metabolic strategies for improved production of Escherichia coli-derived 6-deoxyerythronolide B. Appl Environ Microbiol 68:3287–3292
Pfleger BF, Pitera DJ, Newman JD, Martin VJ, Keasling JD (2007) Microbial sensors for small molecules: development of a mevalonate biosensor. Metab Eng 9:30–38
Prather KL, Martin CH (2008) De novo biosynthetic pathways: rational design of microbial chemical factories. Curr Opin Biotechnol 19:468–474
Price LA, Kajkowski EM, Hadcock JR, Ozenberger BA, Pausch MH (1995) Functional coupling of a mammalian somatostatin receptor to the yeast pheromone response pathway. Mol Cell Biol 15:6188–6195
Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, Chang MC, Withers ST, Shiba Y, Sarpong R, Keasling JD (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440:940–943
Roth RJ, Acton N (1989) A simple conversion of artemisinic acid into artemisinin. J Nat Prod 52:1183–1185
Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D (2008) Kemp elimination catalysts by computational enzyme design. Nature 453:190–195
Rude MA, Khosla C (2006) Production of ansamycin polyketide precursors in Escherichia coli. J Antibiot (Tokyo) 59:464–470
Sachpatzidis A, Benton BK, Manfredi JP, Wang H, Hamilton A, Dohlman HG, Lolis E (2003) Identification of allosteric peptide agonists of CXCR4. J Biol Chem 278:896–907
Schultz CL, Moini M (2003) Analysis of underivatized amino acids and their D/L-enantiomers by sheathless capillary electrophoresis/electrospray ionization mass spectrometry. Anal Chem 75:1508–1513
Scott CP, Abel-Santos E, Wall M, Wahnon DC, Benkovic SJ (1999) Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci USA 96:13638–13643
Serrano L (2007) Synthetic biology: promises and challenges. Mol Syst Biol 3:158
Suess B, Hanson S, Berens C, Fink B, Schroeder R, Hillen W (2003) Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res 31:1853–1858
Suess B, Fink B, Berens C, Stentz R, Hillen W (2004) A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res 32:1610–1614
Tan RX, Lu H, Wolfender JL, Yu TT, Zheng WF, Yang L, Gafner S, Hostettmann K (1999) Mono- and sesquiterpenes and antifungal constituents from Artemisia species. Planta Med 65:64–67
Tang Y, Kim CY, Mathews II, Cane DE, Khosla C (2006) The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc Natl Acad Sci USA 103:11124–11129
Tavassoli A, Benkovic SJ (2005) Genetically selected cyclic-peptide inhibitors of AICAR transformylase homodimerization. Angew Chem Int Ed Engl 44:2760–2763
Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN (2008) Inhibition of HIV budding by a genetically selected cyclic peptide targeting the Gag-TSG101 interaction. ACS Chem Biol 3:757–764
Topp S, Gallivan JP (2010) Emerging applications of riboswitches in chemical biology. ACS Chem Biol 5:139–148
Voigt CA (2006) Genetic parts to program bacteria. Curr Opin Biotechnol 17:548–557
Walker K, Croteau R (2001) Taxol biosynthetic genes. Phytochemistry 58:1–7
Wang L, Brock A, Herberich B, Schultz PG (2001) Expanding the genetic code of Escherichia coli. Science 292:498–500
Wang K, Neumann H, Peak-Chew SY, Chin JW (2007) Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol 25:770–777
Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM (2009) Programming cells by multiplex genome engineering and accelerated evolution. Nature 460:894–898
Watanabe K, Rude MA, Walsh CT, Khosla C (2003a) Engineered biosynthesis of an ansamycin polyketide precursor in Escherichia coli. Proc Natl Acad Sci USA 100:9774–9778
Watanabe K, Wang CC, Boddy CN, Cane DE, Khosla C (2003b) Understanding substrate specificity of polyketide synthase modules by generating hybrid multimodular synthases. J Biol Chem 278:42020–42026
Werstuck G, Green MR (1998) Controlling gene expression in living cells through small molecule–RNA interactions. Science 282:296–298
Widmaier DM, Tullman-Ercek D, Mirsky EA, Hill R, Govindarajan S, Minshull J, Voigt CA (2009) Engineering the Salmonella type III secretion system to export spider silk monomers. Mol Syst Biol 5:309
Xie J, Schultz PG (2006) A chemical toolkit for proteins—an expanded genetic code. Nat Rev Mol Cell Biol 7:775–782
Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM (2008) Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat Rev Microbiol 6:681–691
Yin J, Straight PD, Hrvatin S, Dorrestein PC, Bumpus SB, Jao C, Kelleher NL, Kolter R, Walsh CT (2007) Genome-wide high-throughput mining of natural-product biosynthetic gene clusters by phage display. Chem Biol 14:303–312
Yoshikuni Y, Ferrin TE, Keasling JD (2006) Designed divergent evolution of enzyme function. Nature 440:1078–1082
You L, Cox RS 3rd, Weiss R, Arnold FH (2004) Programmed population control by cell–cell communication and regulated killing. Nature 428:868–871
Zhang W, Li Y, Tang Y (2008) Engineered biosynthesis of bacterial aromatic polyketides in Escherichia coli. Proc Natl Acad Sci U S A 105:20683–20688
Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics 8:215–225
Acknowledgement
The authors gratefully acknowledge the financial support by the German Initiative of Excellence and the Emmy-Noether Program of the German Research Foundation.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Neumann, H., Neumann-Staubitz, P. Synthetic biology approaches in drug discovery and pharmaceutical biotechnology. Appl Microbiol Biotechnol 87, 75–86 (2010). https://doi.org/10.1007/s00253-010-2578-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-010-2578-3