Abstract
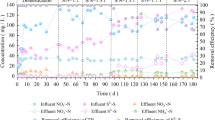
Denitrifying sulfide removal (DSR) process simultaneously converts sulfide, nitrate, and chemical oxygen demand from industrial wastewaters to elemental sulfur, nitrogen gas, and carbon dioxide, respectively. This investigation utilizes a dilution-to-extinction approach at 10−2 to 10−6 dilutions to elucidate the correlation between the composition of the microbial community and the DSR performance. In the original suspension and in 10−2 dilution, the strains Stenotrophomonas sp., Thauera sp., and Azoarcus sp. are the heterotrophic denitrifiers and the strains Paracoccus sp. and Pseudomonas sp. are the sulfide-oxidizing denitrifers. The 10−4 dilution is identified as the functional consortium for the present DSR system, which comprises two functional strains, Stenotrophomonas sp. strain Paracoccus sp. At 10−6 dilution, all DSR performance was lost. The functions of the constituent cells in the DSR granules were discussed based on data obtained using the dilution-to-extinction approach.





Similar content being viewed by others
References
Adav SS, Lee DJ, Lai JY (2009) Functional consortium from aerobic granules under high organic loading rates. Bioresour Technol 100:2546–2550
APHA (1995) Standard methods for the examination of water and wastewater, 20th edn. American Public Health Association, Washington, DC
Assih EA, Ouattara AS, Thierry S, Cayol JL, Labat M, Macarie H (2002) Stenotrophomonas acidaminiphila sp.nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int J Syst Evol Microbiol 52:559–568
Bahambhani Y, Singh M (1991) Physiological effects of hydrogen sulfide inhalation during exercise in healthy men. J Appl Physiol 71:1872–1877
Cadena F, Peters RW (1988) Evaluation of chemical oxidizer for hydrogen sulfide control. J Water Pollut Control Fed 60:1259–1263
Chen C, Ren NQ, Wang AJ, Yu ZG, Lee DJ (2008a) Simultaneous biological removal of sulfur, nitrogen and carbon using EGSB reactor. Appl Microbiol Biotechnol 78:1057–1063
Chen C, Ren NQ, Wang AJ, Lee DJ (2008b) Microbial community of granules in expanded granular sludge bed reactor for simultaneous biological removal of sulfate, nitrate and lactate. Appl Microbiol Biotechnol 79:1071–1077
Chen C, Wang AJ, Ren NQ, Kan HJ, Lee DJ (2008c) Biological breakdown of denitrifying sulfide removal process in high-rate expanded granular bed reactor. Appl Microbiol Biotechnol 81:765–770
Chen C, Wang AJ, Ren NQ, Lee DJ, Lai JY (2009) High-rate denitrifying sulfide removal process in expanded granular sludge bed reactor. Bioresour Technol 100:2316–2319
Gutierrez-Correa M, Porial L, Moreno P, Tengerdy RP (1999) Mixed culture solid substrate fermentation of Trichoderma reesei with Aspergillus niger on sugarcane bagasse. Bioresour Technol 68:173–178
Lipski A, Reichert K, Reuter B, Sproer AK (1998) Identification of bacterial isolates from biofilters as Paracoccus alkenifer sp. nov. and Paracoccus solventivorans with emended description of Paracoccus solventivorans. Int J Syst Bacteriol 48:529–536
Mechichi T, Stackebrandt E, Gadon N, Fuchs G (2002) Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch Microbiol 178:26–35
Reyes-Avila JS, Razo-Flores E, Gomez J (2004) Simultaneous biological removal of nitrogen, carbon and sulfur by denitrification. Water Res 38:3313–3321
Siller H, Rainey FA, Stackebrandt E, Winter J (1996) Isolation and characterization of a new gram-negative acetone-degrading, nitrate-reduxing bacterium from soil, Paracoccus solventivorans sp. nov. Int J Syst Bacteriol 46:1125–1130
Song B, Haggblom MM, Zhou J, Tiedje JM, Palleroni NJ (1999) Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp. nov. and Azoarcus toluclasticus sp. nov. Int J Syst Bacteriol 49:1129–1140
Sorokin DY (2003) Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72:641–653
Sorokin DY, Teske A, Robertson LA, Kuenen JG (1999) Anaerobic oxidation of thiosulfate to tetrathionate by obligately heterotrophic bacteria, belonging to the Pseudomonas stutzeri group. FEMS Microbiol Ecol 30:113–123
Springer N, Ludwig W, Philipp B, Schink B (1998) Azoarcus anaerobius sp. nov., a resorcinol-degrading, strictly anaerobic, denitrifying bacterium. Int J Syst Bacteriol 48:953–956
Sun M, Mu ZX, Chen YP, Sheng GP, Liu XW, Chen YZ, Zhao Y, Wang HL, Yu HQ, Wei L, Mang F (2009) Microbe-assisted sulfide oxidation in the anode of a microbial fuel cell. Environ Sci Technol 43:3372–3377
Tajima K, Aminov RI, Nagamine T, Ogata K, Nakamura M, Matsui H, Benno Y (1999) Rumen bacterial diversity as determined by sequence analysis of 16SrDNA libraries. FEMS Microbiol Ecol 29:159–169
Tengerdy KH (1996) Cellulase production by solid-state fermentation. J Sci Ind Res 55:313–316
Truper HG, Schlegel HG (1964) Sulfur metabolism of Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie Van Leeuwenhoek 30:225–38
Wang AJ, Gao LF, Ren NQ, Xu JF, Liu C, Lee DJ (2009) Enrichment strategy to select functional consortium from mixed cultures. Int J Hydrogen Energy (in press)
Watanabe K, Kodama Y, Harayama S (2001) Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J Microbiol Methods 44:253–262
Weimer PJ (1991) Use of mixed cultures for the production of commercial chemicals. In: Zeikus G, Johnson EA (eds) Mixed cultures in biotechnology. McGraw-Hill, New York, pp 205–232
Zhou J, Fries MR, Chee-Sanford JC, Tiedje JM (1995) Phylogenetic analyses of a new group of denitrifiers capable of anaerobic growth of toluene and description of Azoarcus tolulyticus sp. nov. Int J Syst Bacteriol 45:500–506
Acknowledgement
This project is supported by the State Key Laboratory of Water Resource and Environment (SKLWRE), Harbin Institute of Technology (HIT), China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, C., Ren, N., Wang, A. et al. Functional consortium for denitrifying sulfide removal process. Appl Microbiol Biotechnol 86, 353–358 (2010). https://doi.org/10.1007/s00253-009-2367-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2367-z