Abstract
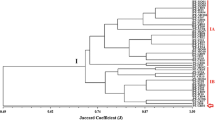
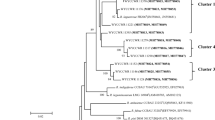
In tropical soils, diversity and biotechnological potential of symbiotic diazotrophic bacteria are high. However, the phylogenetic relationships of prominent strains are still poorly understood. In addition, in countries such as Brazil, despite the broad use of rhizobial inoculants, molecular methods are rarely used in the analysis of strains or determination of inoculant performance. In this study, both rep-PCR (BOX) fingerprintings and the DNA sequences of the 16S rRNA gene were obtained for 54 rhizobial strains officially authorized for the production of commercial inoculants in Brazil. BOX-PCR has proven to be a reliable fingerprinting tool, reinforcing the suggestion of its applicability to track rhizobial strains in culture collections and for quality control of commercial inoculants. On the other hand, the method is not adequate for grouping or defining species or even genera. Nine strains differed in more than 1.03% (15) nucleotides of the 16S rRNA gene in relation to the closest type strain, strongly indicative of new species. Those strains were distributed across the genera Burkholderia, Rhizobium, and Bradyrhizobium.


Similar content being viewed by others
References
Alberton O, Kaschuk G, Hungria M (2006) Sampling effects on the assessment of genetic diversity of rhizobia associated with soybean and common bean. Soil Biol Biochem 38:1298–1307
Allen ON, Allen E (1981) The Leguminosae: A source book of characteristics, uses and nodulation. The University of Wisconsin Press, Madison
Barcellos FG, Menna P, Batista JSS, Hungria M (2007) Evidence of horizontal transfer of symbiotic genes from a Bradyrhizobium japonicum inoculant strain to indigenous Sinorhizobium (Ensifer) fredii and Bradyrhizobium elkanii in a Brazilian savannah soil. Appl Environ Microb 73:2635–2643
Batista JSS, Hungria M, Barcellos FG, Ferreira MC, Mendes IC (2007) Variability in Bradyrhizobium japonicum and B. elkanii seven years after introduction of both the exotic microsymbiont and the soybean host in a cerrados soil. Microb Ecol 53:270–284
de Bruijn FJ (1992) Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacterial. Appl Environ Microb 58:2180–2187
Doignon-Bourcier F, Sy A, Willems A, Torck U, Dreyfus B, Gillis M, de Lajudie P (1999) Diversity of bradyrhizobia from 27 tropical Leguminosae species native of Senegal. Syst Appl Microbiol 22:647–661
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
FEPAGRO (Fundação Estadual de Pesquisa Agropecuária) (1999) Culture Collection Catalogue, 8th edn. FEPAGRO, Porto Alegre
Germano MG, Menna P, Mostasso FL, Hungria M (2006) RFLP analysis of the rRNA operon of a Brasilian collection of bradyhizobial strains from 33 legumes species. Int J Syst Evol Micr 56:217–229
Grange L, Hungria M (2004) Genetic diversity of indigenous common bean (Phaseolus vulgaris) rhizobia in two Brazilian ecosystems. Soil Biol Biochem 36:1389–1398
Hedges SB (1992) The number of replications needed for accurate estimation of the bootstrap p-value in phylogenetic studies. Mol Biol Evol 9:366–369
Hungria M, Campo RJ (2007) Inoculantes microbianos: situação no Brasil. In: Izaguirre-Mayoral ML, Labandera C, Sanjuan J (eds) Biofertilizantes en Iberoamérica: Visión Técnica, Científica y Empresarial. Cyted/Biofag, Montevideo, pp 22–31
Hungria M, Franchini JC, Campo RJ, Graham PH (2005) The importance of nitrogen fixation to soybean cropping in South America. In: Werner W, Newton WE (eds) Nitrogen fixation in agriculture, forestry, ecology and the environment. Springer, Dordrecht Amsterdam, pp 25–42
Hungria M, Chueire LMO, Megías M, Lamrabet Y, Probanza A, Guttierrez-Mañero FJ, Campo RJ (2006a) Genetic diversity of indigenous tropical fast-growing rhizobia isolated from soybean nodules. Plant Soil 288:343–356
Hungria M, Campo RJ, Mendes IC, Graham PH (2006b) Contribution of biological nitrogen fixation to the N nutrition of grain crops in the tropics: the success of soybean (Glycine max L. Merr.) in South America. In: Singh RP, Shankar N, Jaiwal PK (eds) Nitrogen nutrition and sustainable plant productivity. Studium Press LLC, Houston, pp 43–93
ILDIS (International Legume Database & Information Service) (2005) Retrieved April 28th. http://www.ildis.org
Jaccard P (1912) The distribution of flora in the alpine zone. New Phytol 11:37–50
Kaschuk G, Hungria M, Andrade DS, Campo RJ (2006) Genetic diversity of rhizobia associated with common bean (Phaseolus vulgaris L.) grown under no-tillage and conventional systems in Southern Brazil. Appl Soil Ecol 32:210–220
Kim W, Song M, Song W, Kim K, Chung S, Choi C, Park Y (2003) Comparison of 16 S rDNA analysis and rep-PCR genomic fingerprinting for molecular identification of Yersinia pseudotuberculosis. Ant van Leeuwenhoek 83:125–133
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Laguerre G, van Berkum P, Amarger N, Prevost D (1997) Genetic diversity of rhizobial symbionts isolated from legume species within the genera Astragalus, Oxytropis, and Onobrychis. Appl Environ Microb 63:4748–4758
MAPA (Ministério da Agricultura, Pecuária e Abastecimento) (2006) Instrução normativa N 10, Retrieved March 21th. http://extranet.agricultura.gov.br/sislegis-consulta/consultarLegislacao.do?operacao=visualizar&id=16735
Menna P, Hungria M, Barcellos FG, Bangel EV, Hess PN, Martinez-Romero E (2006) Molecular phylogeny based on the 16S rRNA gene of elite rhizobial strains used in Brazilian commercial inoculants. Syst Appl Microbiol 29:315–332
Menna P, Pereira AA, Bangel EV, Hungria M (2009) rep-PCR of tropical rhizobia for strain fingerprinting, biodiversity appraisal and as a taxonomic and phylogenetic tool. Symbiosis 48 (1-3) in press
Mostasso L, Mostasso FL, Dias BG, Vargas MAT, Hungria M (2002) Selection of bean (Phaseolus vulgaris L.) rhizobial strains for the Brazilian Cerrados. Field Crop Res 73:121–132
Oyaizu H, Naruhashi N, Gamou T (1992) Molecular methods of analysing bacterial diversity: The case of rhizobia. Biodivers Conserv 1:237–249
Pinto FGS, Hungria M, Mercante FM (2007) Polyphasic characterization of Brazilian Rhizobium tropici strains effective in fixing N2 with common bean (Phaseolus vulgaris L.). Soil Biol Biochem 39:1851–1864
Polhill RM, Raven PH (1981) Advances in legume systematics. Royal Botanic Gardens, Kew
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sneath PBA, Sokal RR (1973) Numerical taxonomy. WH Freeman & Co, San Francisco
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–7882
Urtz BE, Elkan GH (1996) Genetic diversity among Bradyrhizobium isolates that effectively nodulate peanut (Arachis hypogaea). Can J Microbiol 42:1121–1130
Versalovic J, Schneider M, de Bruijn FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence based PCR (rep-PCR). Meth Mol Cell Biol 5:25–40
Vincent JM (1970) Manual for the practical study of root nodule bacteria. Blackwell, Oxford
Vinuesa P, Rademaker JLW, de Bruijn FJ, Werner D (1998) Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-Restriction Fragment Length Polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16 S–23S rRNA intergenic spacers, repetitive extragenic palindomic PCR genomic fingerprinting, and partial 16S rRNA sequencing. Appl Environ Microbiol 64:2096–2104
Vinuesa P, Leon-Barrios M, Silva C, Willems A, Jarabo-Lorenzo A, Perez-Galdona R, Werner D, Martinez-Romero E (2005) Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int J Syst Evol Microbiol 55:569–575
Acknowledgments
The work was partially supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil), projects 552393/2005-3 and 577933/2008-6, PQ (300698/2007), and PNPD (558455/2008-5). The authors thank also Ligia Maria Oliveira Chueire (Embrapa Soja) for help in several steps of this work and Dr. Allan R.J. Eaglesham for helpful discussion. P. Menna acknowledges fellowships from CNPq (projects 505946/2004-1 and 558455/2008-5), and F.G. Barcellos acknowledges a pro-doc fellowship from CAPES.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Binde, D.R., Menna, P., Bangel, E.V. et al. rep-PCR fingerprinting and taxonomy based on the sequencing of the 16S rRNA gene of 54 elite commercial rhizobial strains. Appl Microbiol Biotechnol 83, 897–908 (2009). https://doi.org/10.1007/s00253-009-1927-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-1927-6