Abstract
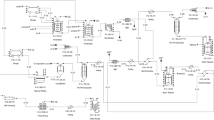
The aim of the present study was to design an in vitro model system to evaluate the probiotic potential of food. A single bioreactor system—gastrointestinal tract simulator (GITS) was chosen for process simulation on account of its considerable simplicity compared to multi-vessel systems used in previous studies. The bioreactor was evaluated by studying the viability of four known probiotic bacteria (Lactobacillus acidophilus La-5, Lactobacillus johnsonii NCC 533, Lactobacillus casei strain Shirota, and Lactobacillus rhamnosus GG) as a function of their physiological state. L. acidophilus and L. johnsonii survived in GITS better when introduced at an early stationary or exponential phase compared to being previously stored for 2 weeks at 4 °C. These two species were more resistant to bile salts and survived better than L. casei and L. rhamnosus GG. The latter two species gave large losses (up to 6 log) in plate counts independent of growth state due to the bile. However, experiments with some commercial probiotic products containing Lb. GG bacteria showed much better survival compared with model food (modified deMan–Rogosa–Sharpe growth medium), thus demonstrating the influence of the food matrix on the viability of bacteria. The study demonstrated that GITS can be successfully used for evaluation of viability of probiotic bacteria and functionality of probiotic food.


Similar content being viewed by others
References
Alander M, Satokari R, Korpela R, Saxelin M, Vilpponen-Salmela T, Mattila-Sandholm T, von Wright A (1999a) Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl Environ Microbiol 65:351–354
Alander M, De Smet I, Nollet L, Verstraete W, von Wright A, Mattila-Sandholm T (1999b) The effect of probiotic strains on the microbiota of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME). Int J Food Microbiol 46:71–79
Begley M, Gahan CGM, Hill C (2005) The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651
Charteris WP, Kelly PM, Morelli L, Collins JK (1998) Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbiol 84:759–768
De Man JC, Rogosa M, Sharpe ME (1960) A medium for the cultivation of lactobacilli. J Appl Bacteriol 23:130–135
Ewe K, Karbach U (1990) Gastrointestinal tract. In: Schmidt RF, Thews G (eds) Physiologie des Menschen. 24th edn. Springer, Berlin, pp 733–777
Fonden R, Bjorneholm S, Ohlson K (2000) Lactobacillus F19—a new probiotic strain. In: Poster presented at the fourth workshop of the PROBDEMO-FAIR CT 96-1028 project Functional Foods for EU health in 2000
Gmeiner M, Kneifel W, Kulbe KD, Wouters R, De Boever P, Nollet L, Verstraete W (2000) Influence of a synbiotic mixture consisting of Lactobacillus acidophilus 74-2 and a fructooligosaccharide preparation on the microbial ecology sustained in a simulation of the human intestinal microbial ecosystem (SHIME reactor). Appl Microbiol Biotechnol 53:219–223
Goldin BR, Gorbach SL, Saxelin M, Barakat S, Gualtieri L, Salminen S (1992) Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci 37:121–128
Hyronimus B, Le Marrec C, Hadj Sassi A, Deschamps A (2000) Acid and bile tolerance of spore-forming lactic acid bacteria. Int J Food Microbiol 61:193–197
Jacobsen CN, Rosenfeldt Nielsen V, Hayford AE, Møller PL, Mickaelsen KF, Pærregaard A, Sandström B, Tvede M, Jakobsen M (1999) Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl Environ Microbiol 65:4949–4956
Kontula P, Jaskari J, Nollet L, De Smet I, von Wright A, Poutanen K, Mattila-Sandholm T (1998) The colonization of a simulator of the human intestinal microbial ecosystem by a probiotic strain fed on a fermented oat bran product: effects on the gastrointestinal microbiota. Appl Microbiol Biotechnol 50:246–252
Kos B, Šušković JJG, Matošić S (2000) Effect of protectors on the viability of Lactobacillus acidophilus M92 in simulated gastrointestinal conditions. Food technol biotechnol 38:121–127
Kristoffersen SM, Ravnum S, Tourasse NJ, Økstad OA, Kolstø AB, Davies W (2007) Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14570. J Bacteriol 189:5302–5313
Lloyd D, Hayes AJ (1995) Vigour, vitality and viability of microorganisms. FEMS Microbiol Lett 133:1–7
Mainville I, Arcand Y, Farnworth ER (2005) A dynamic model that simulates the human upper gastrointestinal tract for the study of probiotics. Int J Food Microbiol 99:287–296
Marteau P, Pochart P, Bouhnik Y, Zidi S, Goderel I, Rambaud JC (1992) Survie dans l’intestin grêle de Lactobacillus acidophilus et Bifidobacterium spp. ingérés dans un lait fermenté: une base rationnelle pour l’utilisation des probiotiques chez l’homme. Gastroentérol Clin Biol 16:25
Marteau P, Minekus M, Havenaar R, Huis in’t Veld JHJ (1997) Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: validation and the effects of bile. J Dairy Sci 80:1031–1037
Mattila-Sandholm T, Mättö J, Saarela M (1999) Lactic acid bacteria with health claims—interactions and interference with gastrointestinal flora. Int Dairy J 9:25–35
Minekus M, Marteau P, Havenaar R, Huis in’t Veld JHJ (1995) A multicompartmental dynamic computer-controlled model simulating the stomach and the small intestine. Alt Lab Anim 23:197–209
Molly K, Vande Woestyne M, Verstraete W (1993) Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol 39:254–258
Nollet L, Vande Velde I, Verstraete W (1997) Effect of the addition of Peptostreptococcus productus ATCC35244 on the gastro-intestinal microbiota and its activity, as simulated in an in vitro simulator of the human gastrointestinal tract. Appl Microbiol biotechnol 48:99–104
Prasad J, Gill H, Smart J, Gopal PK (1998) Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int Dairy J 8:993–1002
Robertson DS (2005) The chemical reactions in the human stomach and the relationship to metabolic disorders. Med Hypoth 64:1127–1131
Siezen RJ, van Enckevort FH, Kleerebezem M, Teusink B (2004) Genome data mining of lactic acid bacteria: the impact of bioinformatics. Curr Opin Biotechnol 15:105–115
Spanhaak S et al (1998) The effect of consumption of milk fermented by Lactobacillus acidophilus strain Sirota on the intestinal microflora and immune parameters in humans. Eur J Clin Nutr 52:899–907
Acknowledgements
The financial support for this research was provided by the Enterprise Estonia project EU22704, Ministry of Education, Estonia, through the grant SF0142497s03 and Estonian Science Foundation grant G6715. Authors thank NESTEC for providing L. johnsonii NCC533 strain. We are also grateful to David Schryer for revising the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sumeri, I., Arike, L., Adamberg, K. et al. Single bioreactor gastrointestinal tract simulator for study of survival of probiotic bacteria. Appl Microbiol Biotechnol 80, 317–324 (2008). https://doi.org/10.1007/s00253-008-1553-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1553-8