Abstract
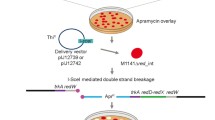
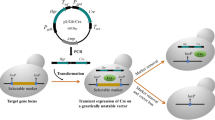
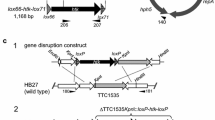
Site-specific recombinases revolutionized “in vivo” genetic engineering because they can catalyze precise excisions, integrations, inversions, or translocations of DNA between their distinct recognition target sites. We have constructed a synthetic gene encoding Cre recombinase with the GC content 67.7% optimized for expression in high-GC bacteria and demonstrated this gene to be functional in Streptomyces lividans. Using the synthetic cre(a) gene, we have removed an apramycin resistance gene flanked by loxP sites from the chromosome of S. lividans with 100% efficiency. Sequencing of the chromosomal DNA part showed that excision of the apramycin cassette by Cre recombinase was specific.


Similar content being viewed by others
References
Bierman M, Logan R, O’Brien K, Seno ET, Rao RN, Schoner BE (1992) plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49
Branda C, Dymecki S (2004) Talking about revolution: the impact of site-specific recombinases on genetic analyses in mice. Develop Cell 6:7–28
Calderone TL, Stevens RD, Oas TG (1996) High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J Mol Biol 262:407–412
Collins EC, Pannell R, Simpson EM, Forster A, Rabbitts TH (2000) Inter-chromosomal recombination of Mll and Af9 genes mediated by cre–loxP in mouse development. EMBO Rep 1:127–132
Chater KF (2000) Developmental decisions during sporulation in the aerial mycelium in Streptomyces. In: Brun YV, Shimkets LJ (eds) Prokaryotic development. American Society for Microbiology Press, Washington, DC, pp 33–48
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645
Gopaul DN, Guo F, Duyne GDV (1998) Structure of the Holliday junction intermediate in Cre/loxP site-specific recombination. EMBO J 17:4175–4187
Gust B, Challis GL, Fowler K, Kieser T, Chater KF (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546
Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22:346–353
Hesketh A, Bucca G, Laing E, Flett F, Hotchkiss G, Smith CP, Chater KF (2007) New pleiotropic effects of eliminating a rare tRNA from Streptomyces coelicolor, revealed by combined proteomic and transcriptomic analysis of liquid cultures. BMC Genomics 8:261–283
Hanahan D (1983) Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166:557–580
Kane JF (1995) Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol 6:494–500
Khodakaramian G, Lissenden S, Gust B, Moir L, Hoskisson PA, Chater K et al (2006) Expression of Cre recombinase during transient phage infection permits efficient marker removal in Streptomyces. Nucleic Acids Res 34:e20
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood D (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Kobler L, Schwertfirm G, Schmieger H, Bolotin A, Sladkova I (1991) Construction and transduction of a shuttle vector bearing the cos site of Streptomyces phage jC31 and determination of its cohesive ends. FEMS Microbiol Lett 62:347–353
Luzhetskyy A, Pelzer S, Bechthold A (2007) The future of natural products as a source of new antibiotics. Curr Opin Investig Drugs 8:608–613
Luzhetskyy A, Zhu L, Gibson M, Fedoryshyn M, Dürr C, Hofmann C, Hoffmeister D, Ostash B, Mattingly C, Adams V, Fedorenko V, Rohr J, Bechthold A (2005) Generation of novel landomycins M and O through targeted gene disruption. Chembiochem 6:675–678
Luzhetskyy A, Fedoryshyn M, Gromyko O, Ostash B, Rebets Y, Bechthold A, Fedorenko V (2006) IncP plasmids are most effective in mediating conjugation between Escherichia coli and streptomycetes. Genetika 42:595–601
O'Gorman S, Fox DT, Wahl GM (1991) Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science 251:1351–1355
Ohama T, Muto A, Osawa S (1990) Role of GC-biased mutation pressure on synonymous codon choice in Micrococcus luteus, a bacterium with a high genomic GC-content. Nucleic Acid Res 18:1565–1569
Posfai G, Koob MD, Kirkpatrick HA, Blattner FR (1997) Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol 179:4426–4428
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB (2003) A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol 21:562–565
Schweizer HP (2003) Application of the Saccharomyces cerevisiae Flp–FRT system in bacterial genetics. J Mol Microbiol Biotechnol 5:67–77
Slauch JM, Camilli A (2000) IVET and RIVET: use of gene fusions to identify bacterial virulence factors specifically induced in host tissues. Methods Enzymol 326:73–96
Stark WM, Boocock MR, Sherratt DJ (1992) Catalysis by site-specific recombinases. Trends Genet 8:432–439
Voeykova T, Slavinskaya E, Orekhov A, Lomovskaya N (1979) Identification of restriction and modification systems in Streptomyces strains. Genetika 15:1746–1756
Zhang Y, Buchholz F, Muyrers JP, Stewart AF (1998) A new logic for DNA engineering using recombination in Escherichia coli. Nature Genet 20:123–128
Acknowledgment
The authors wish to thank Dr. B. Gust for providing plasmid vector pIJ774. This work was supported by a grant from the BMBF (GenoMic-plus) to A.B. and by a grant from Baden-Württemberg to A.L.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fedoryshyn, M., Welle, E., Bechthold, A. et al. Functional expression of the Cre recombinase in actinomycetes. Appl Microbiol Biotechnol 78, 1065–1070 (2008). https://doi.org/10.1007/s00253-008-1382-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-008-1382-9