Abstract
A gene encoding a putrescine oxidase (PuORh, EC 1.4.3.10) was identified from the genome of Rhodococcus erythropolis NCIMB 11540. The gene was cloned in the pBAD vector and overexpressed at high levels in Escherichia coli. The purified enzyme was shown to be a soluble dimeric flavoprotein consisting of subunits of 50 kDa and contains non-covalently bound flavin adenine dinucleotide as a cofactor. From all substrates, the highest catalytic efficiency was found with putrescine (K M = 8.2 μM, k cat = 26 s−1). PuORh accepts longer polyamines, while short diamines and monoamines strongly inhibit activity. PuORh is a reasonably thermostable enzyme with t 1/2 at 50°C of 2 h. Based on the crystal structure of human monoamine oxidase B, we constructed a model structure of PuORh, which hinted to a crucial role of Glu324 for substrate binding. Mutation of this residue resulted in a drastic drop (five orders of magnitude) in catalytic efficiency. Interestingly, the mutant enzyme showed activity with monoamines, which are not accepted by wt-PuORh.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flavoprotein oxidases catalyze the oxidation of a wide range of compounds, while at the same time they reduce oxygen to hydrogen peroxide. They are valuable biocatalysts for the oxidative activation of biomolecules, as they usually selectively oxidize their substrate at a specific position, leaving other positions unaffected. Due to their ability to use molecular oxygen as electron acceptor, no expensive coenzymes like NAD(P)H are needed. This makes oxidases inexpensive and rather straightforward in usage compared to other redox enzymes.
By far, the best studied oxidase is glucose oxidase from Aspergillus niger. This enzyme has been applied for decades, mostly for diagnostic applications (Wilson and Turner 1992). Another example is pyranose oxidase (glucose-2-oxidase) from Peniophora gigantea, which oxidizes specifically the C2 position of glucose and has been applied for the chemoenzymatic synthesis of rare sugars and sugar-based synthons (Giffhorn et al. 2000). Human monoamine oxidases (MAO-A and MAO-B) are important for the oxidation of neurotransmitters (Abell and Kwan 2001). An engineered mutant of the homologous A. niger enzyme (MAO-N) has been applied in an elegant biocatalytic deracemization process to obtain enantiomerically pure chiral amines (Carr et al. 2005; Dunsmore et al. 2006).
So far, the most described oxidases are from eukaryotic origin, and heterologous expression of the recombinant protein in Escherichia coli for large scale enzyme production can be problematic. In a search for novel bacterial oxidases, we looked in sequenced genomes and identified genes encoding novel oxidases. By this, we discovered oxidases primarily acting on alditols (Heuts et al. 2007) and phenolic compounds (Jin et al. 2007). These oxidases, both from actinomycetes, could be highly overexpressed in E. coli. Another way of searching for novel bacterial oxidases is to screen unsequenced bacterial genome libraries directly for genes encoding enzyme activity. As actinomycetes appear to be rich in oxidases, we have chosen to explore these organisms for relevant oxidases. Using a plate-based screening protocol for oxidase activity (Alexeeva et al. 2002), we screened a genomic DNA library of the actinomycete Rhodococcus erythropolis NCIMB 11540 to find novel oxidases acting on polyols and/or amines.
In this paper, we describe the discovery and characterization of a novel flavin-containing oxidase primarily acting on putrescine. This putrescine oxidase (PuORh) shares 67% sequence identity with the enzyme from Micrococcus rubens (PuOMr; Ishizuka et al. 1993). We show that PuORh is highly overexpressed in E. coli and can be easily purified. Using a structural model, we were able to identify a glutamate residue (Glu324) that is crucial for substrate binding, and we could alter the substrate specificity by protein engineering.
Materials and methods
Chemicals
Restriction enzymes were from Roche and New England Biolabs. One-shot electrocompetent E. coli TOP10 cells and the TOPO TA Cloning Kit were purchased from Invitrogen. Plasmid isolation was performed using the Qiagen DNA purification kit. Oligonucleotides were purchased from Sigma. Nitrocellulose filters (Protran BA85 132 mm, 0.45 μm pore size) were from Schleicher and Schuell BioScience, Dassel (Germany). Deprenyl and rasagiline were a kind gift from Prof. A. Mattevi (University of Pavia, Italy). All other chemicals were of analytical grade. The genomic DNA library from R. erythropolis NCIMB 11540 was provided by DSM (Geleen, The Netherlands). Constructs were sequenced at GATC Biotech (Kostanz, Germany).
Plate-based screening method for oxidase activity
A gene library of R. erythropolis NCIMB 11540 in pZErO-2 was screened for oxidases using the plate-based oxidase activity screening method adapted from the group of Turner (Alexeeva et al. 2002). This gene library was constructed by partial digestion of genomic DNA from R. erythropolis NCIMB 11540 by Sau3A1. Fragments of 4–10 kb were isolated, ligated in pZErO-2 (cut with BamHI), and followed by transformation of E. coli DH10B. Colonies were collected from plate, stored as glycerol stock, and plasmids were isolated. The total amount of plasmids contained a mean insert size of 6.0 kb and 1% of self-ligated vector molecules.
Electrocompetent E. coli TOP10 cells were transformed with the gene library, and the transformed cells were diluted in Luria–Bertani (LB) medium to obtain single colonies on plate. The diluted cell suspensions were plated on nitrocellulose filters. The filters were placed on top of LB agar containing 0.05 mg/ml kanamycin and incubated for 48 h at 30°C. Subsequently, the nitrocellulose filters were transferred to empty petri dishes and stored at −20°C to partially lyse the cells. Each filter was submersed with 50 ml 50 mM sodium phosphate buffer pH 7.5 containing 1% (w/v) agarose, 2 U/ml Horseradish peroxidase (HRP) and a mixture of 1 mM 4-chloro-1-naphthol, 100 μM cholesterol, 10 mM xylitol, 10 mM sarcosine, 10 mM l-alanine, 10 mM d-glucose, 10 mM d-galactose, 10 mM lactose, and 10 mM d-glucosamine. Plates were incubated at room temperature and regularly checked by visual inspection for color formation in and around colonies. Positives were picked and cultivated overnight in 5 ml liquid LB medium and subjected to another screening cycle to assure that color formation was caused by single clones.
Sequence analysis
To identify unique positive clones, plasmids were isolated and subjected to restriction analysis by EcoRI. Inserts were sequenced and open reading frames (ORFs) were identified using the ORF Finder tool of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). A BLAST search (blastp) was performed with the protein sequence of PuORh using the BLAST function of the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST).
Cloning and expression of the gene encoding PuORh
To overexpress the novel putrescine oxidase, the corresponding gene (puo Rh) was amplified from the fragment of genomic DNA of R. erythropolis using the following primers: puoRh_fw: 5′-GCTCCATATGCCTACTCTCCAGAGAGATG (NdeI site shown in italics) and puoRh_rv: 5′-GCTCAAGCTTTCAGGCCTTGCTGCGGGCG (HindIII site shown in italics). The amplified gene was purified from gel and ligated between the NdeI and HindIII restriction sites of the pBAD/Myc-His vector (Invitrogen). The plasmid carrying the puo Rh gene (pBADpuo Rh) was transformed to CaCl2-competent E. coli TOP10 cells and spread on LB agar plates containing 50 μg/ml of ampicillin. E. coli TOP10 cells containing pBADpuo Rh were tested for overexpression of the protein at 17, 30, and 37°C and at arabinose concentrations of 0, 0.00002, 0.0002, 0.002, 0.02, and 0.2% (w/v). Cell extracts and cell-free extracts of E. coli TOP10 containing pBADpuo Rh were analyzed on a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel to find the best conditions for overexpression of soluble PuORh.
Enzyme purification
To obtain purified protein, E. coli TOP10 containing pBADpuo Rh was cultivated for 24 h at 30°C in 1 l of terrific broth medium containing 50 μg/ml ampicillin and 0.02% (w/v) arabinose. Cells were harvested by centrifugation for 15 min at 6,000 rpm at 4°C. The supernatant was discarded, and the pellet was suspended in 40 ml of 50 mM Tris–HCl pH 7.5 and sonicated for 10 min to break the cells. The cell extract was centrifuged for 30 min at 15,000 rpm at 4°C to remove the broken cells and obtain cell-free extract. From this cell-free extract PuORh was purified using a Q-Sepharose anion exchange column. Unbound protein fractions were washed from the column with 50 mM Tris–HCl buffer pH 7.5. PuORh was eluted from the column with a 50 mM Tris–HCl buffer pH 7.5 by increasing the KCl concentration. Eluted fractions containing PuORh were combined, concentrated using an Amicon filter, and desalted using a HiPrep 26/10 Desalting Column (Amersham Biosciences).
Enzyme activity assay and determination of steady-state kinetic parameters
PuORh activity was measured at 25°C using a peroxidase-coupled assay containing 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) (ABTS) as the chromogenic substrate (Childs and Bardsley 1975). The H2O2 formed by PuORh can be coupled to the Horseradish-peroxidase-mediated oxidation of ABTS resulting in the formation of a green product that can be measured at 420 nm (ɛ 420 = 35.7 mM−1 cm−1). The standard assay mixture with ABTS (HRP-ABTS assay) contained 50 μM putrescine, 50 mM Tris–HCl buffer, pH 8.0, 100 μM ABTS, 5 U of Horseradish peroxidase, 10 μl of enzyme dilution (appropriately diluted), and a fixed amount of substrate. The reaction was started with adding the enzyme solution. Alternatively, 0.1 mM 4-aminoantipyrine (AAP) and 1.0 mM 3,5-dichloro-2-hydroxybenzenesulfonic acid (DCHBS) were used as chromogenic substrates for HRP (Federico et al. 1997; HRP-AAP/DCHBS assay). The oxidation of these substrates results in the formation of a purple product, which can be measured at 515 nm (ɛ 515 = 26 mM−1 cm−1). For the determination of kinetic parameters, PuORh activity was measured at different substrate concentrations at 25°C using the HRP-ABTS enzyme assay. The kinetic parameters of the mutant enzymes (PuORh Glu324Ala and PuORh Glu324Leu) for putrescine were determined by measuring directly the H2O2 production at 240 nm (ɛ 420 = 43.6 M−1 cm−1). Data were fitted with Origin 7.0 using the Michaelis–Menten equation for enzyme kinetics.
Influence of temperature and pH on enzyme activity and stability
To determine the optimal temperature for PuORh activity, the oxidase activity was measured at temperatures between 20 and 60°C. Before adding the enzyme, the assay mixture was equilibrated for 20 min to assure the right temperature. The influence of temperature on the enzyme stability was determined by incubating 1-ml portions of the enzyme at 4, 30, 37, and 50°C. Twenty-microliter samples were taken, placed on ice after which activity was measured. In both experiments, the activity was determined with the HRP-AAP/DCHBS activity assay using 50 μM putrescine and 0.01 μM PuORh.
The pH optimum for putrescine oxidase was determined by measuring the activity at different pH values at 25°C. The following buffers were used: 50 mM Pipes buffer (pH 6.4–7.2), 50 mM Tris–HCl (pH 7.4–8.8), and 50 mM Ches buffer (pH 8.7–9.6). The enzyme activity was measured using the HRP-AAP/DCHBS assay.
Inhibition experiments
Rasagiline, deprenyl, and cyclopropylamine were tested to probe whether they were able to inhibit PuORh by the formation of a covalent adduct with the flavin adenine dinucleotide (FAD) cofactor, as is the case for monoamine oxidase B (Binda et al. 2005). For 900 μl of a 20 μM PuORh solution, an absorbance scan was recorded from 650–300 nm. After the addition of 100 μl of 1 mM inhibitor (final inhibitor concentration, 100 μM), absorbance spectra were recorded regularly in time to see if addition of the inhibitor would cause an alteration of the typical FAD spectrum of PuORh. For cyclopropylamine, also a final inhibitor concentration of 1 mM was tested as described above.
Butylamine, aminoethanol, ethylenediamine, 1,3-diaminopropane, and (2-aminoethyl)-trimethylammonium were tested as competitive inhibitors of PuORh. For this, the Michaelis constant (K M) of PuORh for putrescine was determined for three different concentrations for each of these amino compounds with the HRP-ABTS assay. Inhibition constants (K I) for each compound were calculated with the following formula:
Where [I] represents the inhibitor concentration, K M,inhibitor the apparent Michaelis constant in presence of the inhibitor and the K I the inhibition constant.
Analytical methods
All absorbance spectra were recorded in 50 mM Tris–HCl pH 8.0 at 25°C on a Perkin Elmer Lambda Bio40 spectrophotometer. From a cuvet containing 5 μM PuORh all oxygen was removed by flushing with argon and an absorbance spectrum was recorded from 650 to 300 nm. After adding 50 μM putrescine, another spectrum was recorded for the reduced enzyme. Reoxidation was monitored by collecting spectra in time after the cuvet was exposed to air. A spectrum of the unfolded enzyme was recorded by adding 0.1% SDS and heating for 5 min at 80°C.
Modeling and mutant construction
Based on the structure of MAO-B in complex with rasagiline (PDB/1S2Q; Binda et al. 2004b), a model of PuORh was made using the CHPmodels 2.0 Server. Mutants were constructed by Quick Change PCR. PuORh Glu324Ala was made by a substitution of the codon GAG for GCG using the following primers: puoRhGlu324Ala_fw, 5′-CGAGGTAGTGCAGGCGGTGTACGACAACACC, and puoRhGlu324Ala_rv, 5′-GGTGTT GTCGTACACCGCCTGCACTACCTCG (mutated codon underlined). For construction of PuORh Glu324, the codon GAG was replaced by CTG using the following primers: puoRhGlu324Leu_fw, 5′-CGAGGTAGTGCAGCTGGTGTACGACAACACC, and puoRhGlu324Leu_rv, 5′- GGTGTTGTCGTACACCAGCTGCACTACCTCG (mutated codon underlined). Both mutants were expressed in E. coli TOP 10 and cultivated and purified in the same way as wild-type PuORh.
Nucleotide sequence accession number
The nucleotide sequence of the PuORh encoding gene (puo Rh) has been submitted to GenBank under accession number EU240877.
Results
Identification of a novel putrescine oxidase
To identify novel oxidases acting on alcohols and/or amines, a genomic library from the actinomycete R. erythropolis NCIMB 11540 was screened using a plate-based oxidase assay. Positive clones, expressing oxidase activity, could be identified by their purple color formation. From 55,000 transformants, 5 were observed to produce a purple color around the colony. Retransformation to E. coli TOP10 cells and repeated screening on plate resulted in three positive clones, which showed oxidase activity. Restriction analysis with EcoRI and PvuI showed a different insert for each clone indicating that these were independent clones. Sequencing of one of these clones revealed an inserted fragment of genomic DNA of 5,311 bp. On this fragment, an ORF was identified which shares 67% sequence identity (at the amino acid level) with putrescine oxidase (EC 1.4.3.10) from M. rubens, a FAD-containing amine oxidase that catalyzes the oxidative deamination of putrescine (Ishizuka et al. 1993). When we tested the cell extract of the clone containing the above-mentioned insert of 5,311 bp, we found that it was indeed active with putrescine. The other two positive clones probably contained the same gene, as for both clones by using the primers puoRh_fw and puoRh_rv a PCR product was seen on gel.
Gene cloning, overexpression, and protein purification
For overexpression, the PuORh encoding gene was cloned into the pBAD/Myc-His A vector behind the araBAD promotor (without His-tag), and the plasmid carrying the puo gene was transformed to E. coli TOP10. Several temperatures (17, 30, and 37°C) and inducing conditions (0–0.2% L-(+)-arabinose) were tested to find the optimal conditions for overexpression. The gene was well expressed at arabinose concentrations of 0.02 and 0.2% as judged by a dominant protein band of ±54 kDa on SDS-PAGE. At 17 and 30°C, the protein was mainly present in the soluble fractions while at 37°C the unsolube fractions contained most of the overexpressed protein. The overexpressed protein did not show any fluorescence under UV-light upon SDS-PAGE, which is an indication that this protein does not contain a covalently histidyl-bound flavin cofactor (Fraaije et al. 1997).
PuORh was purified from a 1-l culture containing 0.02% arabinose that was grown for 24 h at 30°C. Due to the high level of overexpression (200 mg), PuORh could be easily purified in one-step by anion exchange chromotography. SDS-PAGE analysis of the purified protein revealed that, besides the major protein band present at around ∼54 kDa, there is a minor band corresponding to a protein mass which is approximately 3 kDa larger (Fig. 1, lane A). It was concluded that this must be the product of the same gene with a His-tag extension, as this minor protein band could be removed by an additional purification step using a Ni-agarose column. (Fig. 1, lane B). Although in the plasmid pBADpuo Rh the stop codon (TGA) was properly introduced, it is known that in some cases, this stop codon can be translated (MacBeath and Kast 1998). With the constructed plasmid, this would indeed result in a 3 kDa larger protein.
Spectral and molecular properties of PuORh
PuORh shows a typical flavoprotein spectrum with absorbance maxima at 377 and 459 nm (Fig. 2). In the presence of 5% trichloroacetic acid, PuORh precipitated and was visible as a white pellet after centrifugation. The supernatant contained a yellow color, and its absorbance spectrum with maxima at 375 and 450 nm was typical for that of free FAD. This shows that the FAD cofactor in PuORh, like in PuOMr, is not covalently bound. At room temperature, the addition of 0.1% SDS did not unfold PuORh, indicating this flavoprotein is rather robust. When 50 μM putrescine was mixed with PuORh under anaerobic conditions, the FAD cofactor was completely reduced (Fig. 2, dotted line). Heating the same sample for 5 min at 80°C completely unfolded the enzyme as can be seen from the resulting free FAD UV/VIS spectrum (Fig. 2, dashed line). The reduced enzyme could be rapidly and fully reoxidized by addition of oxygen, indicating that the enzyme is a true oxidase. From the difference in absorbance between protein-bound FAD and free FAD, the molar extinction coefficient for PuORh (ɛ 459) was calculated, 11.0 mM−1 cm−1. Based on the protein concentration as determined by Wadell’s method (Wolf 1983) and the FAD concentration (A459), a protein/FAD ratio of 0.5 was calculated. This is consistent with what has been described earlier for PuOMr (Desa 1972). Incubation of PuORh with additional FAD did not yield an increase of FAD incorporation or oxidase activity. Gel filtration experiments revealed that PuORh is mainly present as a dimer of ±100 kDa. This corresponds well with the theoretical monomeric mass of 49,375 Da.
Temperature and pH dependence of activity and stability
To investigate the effect of temperature on enzyme activity and stability, both were determined at different temperatures. The optimal temperature for enzyme activity was found to be 30°C. Temperature stability experiments revealed that PuORh is a reasonable thermostable enzyme. At 50°C, half of the activity was lost after 2 h, while at 37°C, such a degree of inactivation was reached only after 1 day. PuORh has a rather sharp pH optimum for activity with putrescine with a peak around pH 8. Below pH 6.4, no significant activity can be detected.
Substrate specificity
Several alcohols, amino alcohols, and amines were tested as substrate, and if they turned out to be converted by PuORh, the steady-state kinetic parameters were determined at pH 8.0. Enzyme activity was found with several aliphatic diamines, amino alcohols, and polyamines. From Table 1, it is clear that putrescine is by far the best substrate. With aromatic amines and amino alcohols, no activity was observed. An increase in carbon chain length of the diamine from 4 to 6 C-atoms resulted in a more than 1,000-fold drop in catalytic efficiency (k cat/K M). For diamines containing 3 or 2 C-atoms and for n-butylamine, no activity could be detected. These amines were found to strongly inhibit the enzyme (Table 2). Apparently, they bind efficiently but are not converted. Clearly, a minimum of two amino groups is required to be a substrate for PuORh, and these amino groups should be preferably 4 C-atoms apart. Polyamines like spermine and spermidine were also converted by PuORh but are poor substrates. Compared to putrescine, 4-amino-1-butanol is also a poor substrate, while 1,4-butanediol is not accepted at all. This again indicates that two amino groups are essential for efficient catalysis, and it confirms that PuORh is a true amine oxidase. Inhibition studies showed that aminoethanol is a very effective competitive inhibitor of PuORh (K I = 1.8 μM). Apparently, this amino alcohol binds very strongly in the active site, while it cannot be oxidized. When we tested 2-hydroxyputrescine, which contains an aminoethanol moiety, we found that this compound is readily oxidized. l-Ornithine was found to be a very poor substrate. Apparently, the presence of a negatively charged and relative bulky carboxylic acid group at the C1 position prevents efficient amine oxidation. Taken together, it can be concluded that PuORh has a narrow substrate specificity and is very selective for putrescine.
Model structure of PuORh reveals a key role for Glu324 in substrate binding
PuORh shares 32% sequence identity with human monoamine oxidase B (MAO-B), which is a 59 kDa FAD-containing protein involved in the oxidation of neurotransmitters and other arylalkylamines like benzylamine and phenylethylamine (Shih et al. 1999). Based on the structure of MAO-B (Binda et al. 2004b), it was possible to construct a model for PuORh. The C-terminal membrane anchor of MAO-B is absent in the resulting model structure of PuORh. This could explain why PuORh is well expressed in the cytosol as a soluble protein in contrast to MAO-B, which is membrane associated (Binda et al. 2004a). In the model, the active site of the PuORh exhibits a similar architecture as that of MAO-B (Fig. 3). The catalytically important “aromatic cage,” present in flavin-containing amine oxidases (Li et al. 2006), is formed by His432 and Tyr395. The other two key amino acid residues, Lys296 and Trp385, involved in non-covalent FAD binding (Binda et al. 2002b) can also be found in the model of PuORh. However, there are also some striking differences. In MAO-B, FAD is covalently attached to Cys397 at the C8α position of the flavin. PuORh has an alanine residue at this position (Ala394), which precludes covalent binding of FAD. Furthermore, the active site of PuORh seems to be narrower than that of MAO-B due to the presence of Trp60 and Met173 in PuORh (Tyr60 and Cys172 in MAO-B). The model of PuORh also shows a glutamic acid residue (Glu324) pointing towards the N5 of the FAD, while MAO-B contains a tyrosine at this position (Tyr326). The presence of such a negatively charged amino acid in the active site has already been predicted for PuOMr several decades ago by Swain and Desa. They suggested the presence of a negatively charged carboxyl group in the active site, as PuOMr was found to be irreversibly inactivated by carbodiimides (Swain and Desa 1976). The presence and position of Glu324 can well explain the difference in substrate specificity between MAO-B and PuORh. PuORh prefers short aliphatic diamines, while MAO-B is active with a range of aromatic monoamines. Probably, Glu324 is involved in binding the protonated amino group of the diamine substrate, while the other amino group is positioned in the “aromatic cage” near the N5 of the flavin. Such a mode of binding for putrescine is consistent with our PuORh model. To confirm this hypothesis, we mutated Glu324 into an alanine (PuORh Glu324Ala) and a leucine residue (PuORh Glu324Leu) and found that the catalytic efficiency for putrescine drops, respectively, 100,000 and 200,000-fold (Table 3).
The distance between Glu324 and the N5 of the isoalloxazine ring of the flavin cofactor (9 Å) may explain the specificity for the specific chain length of the substrate. To be a substrate, a minimal carbon chain length of 4 C-atoms is required. Shorter diamines can bind to Glu324 but cannot reach the flavin and therefore are strong competitive inhibitors of PuORh (Table 2). In case of longer diamines or polyamines, the position of the amino group that is oxidized is less favorable compared to the case of putrescine resulting in a decrease in catalytic efficiency. Aliphatic monoamines like n-butylamine will bind to Glu324 via the amine moiety but cannot be oxidized and therefore are also inhibitors of PuORh. Initial substrate screening using 96-wells plates for the mutants PuORh Glu324Ala and PuORh Glu324Leu revealed that both showed some activity with aromatic amines like phenyl-1-butylamine and with aliphatic monoamines (e.g., k cat/K M for n-butylamine = 0.001 s−1 mM−1 at pH 9.0 for PuORh Glu324Leu). Unfortunately, accurate determination of the kinetic parameters of the mutant enzymes is not straightforward due to their low substrate affinity, which requires the use of high concentrations of amines, causing interference with the peroxidase-based assay.
Discussion
In an attempt to discover novel bacterial oxidases with biocatalytic potential, a putrescine oxidase from R. erythropolis NCIMB 11540 was identified. This putrescine oxidase was discovered by using a plate-based screening method for oxidase activity, which has been used before to screen for improved mutants during directed evolution experiments (Alexeeva et al. 2002). Our study shows that this method is also valuable for the discovery of novel bacterial oxidases of unsequenced genomes. Although the physiological substrate of the discovered putrescine oxidase, 1,4-diaminobutane, was not present during screening, clones expressing this oxidase could still be detected based on their oxidase activity. As polyamines like putrescine, spermine, and spermidine are present in most living cells, including E. coli (Tabor and Tabor 1984), this resulted in the formation of hydrogen peroxide and detection of positive clones.
The physiological role of putrescine oxidase in R. erythropolis is probably related to polyamine degradation (Large 1992), which is supported by the presence of a neighboring gene encoding a putative aldehyde dehydrogenase on the sequenced DNA fragment (data not shown). PuORh displays 67% sequence identity with PuOMr, which is the only bacterial putrescine oxidase that has been characterized so far. A BLAST search with the amino acid sequence of PuORh resulted in a high number of homologous (putative) flavin-containing amine oxidases, which are widely distributed among most kingdoms of life. The seven closest homologs (sequence identity >65%) can all be found in actinomycetes. These sequences all contain the active site glutamate (Glu324), involved in substrate binding, the alanine residue (Ala394), which excludes covalent FAD binding, and the residues, which limit the size of the active site cavity (Trp60 and Met173). Most likely, they represent a clade of orthologous putrescine oxidases, which are not active with monoamines, within the family of (putative) flavin-containing amine oxidases (Supplementary information, ESM 1).
PuORh shares some properties with PuOMr. Both enzymes are soluble dimeric proteins of approximately 100 kDa. Interestingly, both contain only 1 mol of non-covalently bound FAD per mole of dimer, which is rather unique among flavoproteins. The substrate specificity of PuORh is very narrow, like PuOMr, the enzyme is very specific for putrescine. Polyamines are also accepted, but aliphatic monoamines are not converted at all. In general, PuORh appears to have a higher affinity for its substrates than PuOMr (K m for putrescine = 8.2 vs 38 μM).
The narrow substrate specificity of PuOMr for di- and polyamines was already rationalized by the suggestion of the presence of an ‘anionic point’ in the active site. This anionic point, likely due to a carboxylate function, binds one positively charged amino group of the substrate, while another amino group is oxidized (Swain and Desa 1976; Okada et al. 1979). Based on sequence alignment between PuORh and PuOMr and a structural model of PuORh, we can now identify Glu344 as the anionic point in PuOMr. The presence of a carboxylate function as a manner to bind a positively charged amino group is not restricted to putrescine oxidase. In polyamine oxidase (PAO), a negatively charged ‘carboxylate ring’ can be found at one side of its substrate tunnel to guide polyamine substrate molecules into the active site (Binda et al. 1999). In addition, in PAO, two glutamate residues (Glu62 and Glu170) are present in the active site opposite to the N5 of the flavin. However, these residues have been suggested to be protonated (Binda et al. 1999). Moreover, in PAO, Glu62 and Glu170 are in close proximity to a substrate secondary amine group, which will be oxidized, while in PuORh, Glu324 interacts with the (primary) amine group, which will not be oxidized. The amine group to be oxidized in PuORh is placed between His432 and Tyr395, near the N5 of the isoalloxazine ring. This structural feature resembles the “aromatic cage” found in other flavin-containing amine oxidases, like MAO-A (De Colibus et al. 2005), MAO-B (Binda et al. 2002a), and PAO (Binda et al. 1999). For MAO-B, this “aromatic cage” has been shown to play a steric role in substrate binding and in flavin accessibility and helps to increase the substrate amine nucleophilicity (Li et al. 2006).
Polyamines like putrescine, cadaverine, spermine, and spermidine are related to cell growth and differentiation processes. Increased levels in body fluids and tissue occur in cancer patients, and therefore, polyamines represent important cancer markers (Casero and Marton 2007). Polyamines are also used to monitor food freshness, as an increased level can be found in spoiled food due to amino acid degradation (Bardócz 1995). Due to efficient heterologous expression and its catalytic properties, PuORh may develop as a valuable diagnostic enzyme for the detection of low amounts of putrescine and polyamines. Moreover, based on the structural model, enzyme redesign may also allow creation of mutants that can be used for synthetic purposes.
References
Abell CW, Kwan SW (2001) Molecular characterization of monoamine oxidases A and B. Prog Nucleic Acid Res Mol Biol 65:129–156
Alexeeva M, Enright A, Dawson MJ, Mahmoudian M, Turner N (2002) Deracemization of α-methylbenzylamine using an enzyme obtained by in vitro evolution. Angew Chem Int Ed 41:3177–3180
Bardócz S (1995) Polyamines in food and their consequences for food quality and human health. Trends Food Sci Tech 6:341–346
Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A (1999) A 30 Å long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure 7:265–276
Binda C, Newton-Vinson P, Hubálek F, Edmondson DE, Mattevi A (2002a) Structure of human monoamine oxidase B, a drug target for the treatment if neurological disorders. Nat Struct Biol 9:22–26
Binda C, Mattevi A, Edmondson DE (2002b) Structure-function relationships in flavoenzyme dependent amine oxidations. J Biol Chem 277:23973–23976
Binda C, Hubálek F, Li M, Edmondson DE, Mattevi A (2004a) Crystal structure of human monoamine oxidase B, a drug target enzyme monotopically inserted into the mitochondrial outer membrane. FEBS Lett 564:225–228
Binda C, Hubálek F, Li M, Herzig Y, Sterling J, Edmondson DE, Mattevi A (2004b) Crystal structures of monoamine oxidase B in complex with four inhibitors of the N-propargylaminoindan class. J Med Chem 47:1767–1774
Binda C, Hubálek F, Li M, Herzig Y, Sterling J, Edmondson DE, Mattevi A (2005) Binding of rasagiline-related inhibitors to human monoamine oxidases: a kinetic and crystallographic analysis. J Med Chem 48:8148–8154
Carr R, Alexeeva M, Dawson MJ, Gotor-Fernandez V, Humphrey CE, Turner NJ (2005) Directed evolution of an amine oxidase for the preparative deracemisation of cyclic secondary amines. ChemBioChem 6:637–639
Casero RA Jr, Marton LJ (2007) Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov 6:373–390
Childs RE, Bardsley WG (1975) The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethylbenzthiazoline-6-sulphonic acid) as chromogen. Biochem J 145:93–103
De Colibus L, Li M, Binda C, Lustig A, Edmondson DE, Mattevi A (2005) Three-dimensional structure of human monoamine oxidase A (MAO A): relation to the structures of rat MAO A and human MAO B. Proc Natl Acad Sci U S A 102:12684–12689
Desa RJ (1972) Putrescine oxidase from Micrococcus rubens. Purification and properties of the enzyme. J Biol Chem 247:5527–5534
Dunsmore CJ, Carr R, Fleming T, Turner N (2006) A chemoenzymatic route to enantiomerically pure cyclic tertiary amines. J Am Chem Soc 128:2224–2225
Federico R, Angelini R, Ercolini L, Venturini G, Mattevi A, Ascenzi P (1997) Competitive inhibition of swine kidney copper amine oxidase by drugs: amiloride, clonidine, and gabexate mesylate. Biochem Biophys Res Commun 240:150–152
Fraaije MW, Pikkemaat M, Van Berkel W (1997) Enigmatic gratuitous induction of the covalent flavoprotein vanillyl-alcohol oxidase in Penicillium simplicissimum. Appl Environ Microbiol 63:435–439
Giffhorn F, Köpper S, Huwig A, Freimund S (2000) Rare sugars and sugar-based synthons by chemo-enzymatic synthesis. Enzyme Microb Technol 27:734–742
Heuts DPHM, van Hellemond EW, Janssen DB, Fraaije MW (2007) Discovery, characterization and kinetic analysis of an alditol oxidase from Streptomyces coelicolor. J Biol Chem 282:20283–20291
Ishizuka H, Horinouchi S, Beppu T (1993) Putrescine oxidase of Micrococcus rubens: primary structure and Escherichia coli. J Gen Microbiol 139:425–432
Jin J, Mazon HFM, van den Heuvel RHH, Janssen DB, Fraaije MW (2007) Discovery of a eugenol oxidase from Rhodococcus sp. strain RHA1. FEBS J 274:2311–2321
Large PJ (1992) Enzymes and pathways of polyamine breakdown in microorganisms. FEMS Microbiol Rev 88:249–262
Li M, Binda C, Mattevi A, Edmondson DE (2006) Functional role of the “aromatic cage” in human monoamine oxidase B: structures and catalytic properties of Tyr435 mutant proteins. Biochemistry 45:4775–4784
MacBeath G, Kast P (1998) UGA read-through artifacts-when popular gene expression systems need a pATCH. Biotechniques 24:789–794
Okada M, Kawashima S, Imahori K (1979) Substrate specificity and reaction mechanism of putrescine oxidase. J Biochem 86:97–104
Okada M, Kawashima S, Imahori K (1980) Mode of inactivation of putrescine oxidase by 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide or metal ions. J Biochem 88:481–488
Shih JC, Chen K, Ridd MJ (1999) Monoamine oxidase: from genes to behavior. Annu Rev Neurosci 22:197–217
Swain WF, Desa RJ (1976) Mechanism of action of putrescine oxidase. Binding characteristics of the active site of putrescine oxidase from Micrococcus rubens. Biochim Biophys Acta 429:331–341
Tabor CW, Tabor H (1984) Polyamines. Annu Rev Biochem 53:749–790
Wilson R, Turner APF (1992) Glucose oxidase: an ideal enzyme. Biosens Bioelectron 7:165–185
Wolf P (1983) A critical reappraisal of Waddell’s technique for ultraviolet spectrophotometric protein estimation. Anal Biochem 129:145–155
Acknowledgments
We thank T. Sonke from DSM Research (Geleen, The Netherlands) for providing the genomic DNA library of Rhodococus erythropolis NCIMB 11540. This research was financially supported by the Integrated Biosynthesis Organic Synthesis (IBOS) program of The Netherlands Organization for Scientific Research (NWO).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
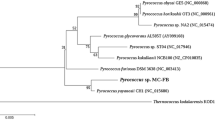
Unrooted phylogenetic tree representation of PuORh homologs, which exhibit over 30% sequence identity (at the amino acid level) throughout the whole sequence. In gray, the clade of putrescine oxidases. MAO-B Human monoamine oxidase B; PuO Rh putrescine oxidase from Rhodococcus erythropolis NCIMB 11540; PuO Mr putrescine oxidase from Micrococcus rubens . (DOC 36 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van Hellemond, E.W., van Dijk, M., Heuts, D.P.H.M. et al. Discovery and characterization of a putrescine oxidase from Rhodococcus erythropolis NCIMB 11540. Appl Microbiol Biotechnol 78, 455–463 (2008). https://doi.org/10.1007/s00253-007-1310-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-007-1310-4