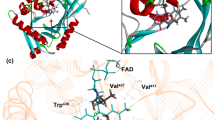
Abstract
A biased mutation-assembling method—that is, a directed evolution strategy to facilitate an optimal accumulation of multiple mutations on the basis of additivity principles, was applied to the directed evolution of water-soluble PQQ glucose dehydrogenase (PQQGDH-B) to reduce its maltose oxidation activity, which can lead to errors in blood glucose determination. Mutations appropriate for the reduction without fatal deterioration of its glucose oxidation activity were developed by an error-prone PCR method coupled with a saturation mutagenesis method. Moreover, two types of incorporation frequency based on their contribution were assigned to the mutations: high (80%) and evens (50%), in constructing a multiple mutant library. The best mutant created showed a marked reduction in maltose oxidation activity, corresponding to 4% of that of the wild-type enzyme, with 35% retention of glucose oxidation activity. In addition, this mutant showed a reduction in galactose oxidation activity corresponding to 5% of that of the wild-type enzyme. In conclusion, we succeeded in developing the PQQGDH-B mutants with improved substrate specificity and validated our method coupled with optimized mutations and their contribution-based incorporation frequencies by applying it to the development.






Similar content being viewed by others
References
Cleton-Jansen AM, Goosen N, Vink K, van de PP (1989) Cloning, characterization and DNA sequencing of the gene encoding the Mr 50,000 quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus. Mol Gen Genet 217:430–436
D’Costa EJ, Higgins IJ, Turner AP (1986) Quinoprotein glucose dehydrogenase and its application in an amperometric glucose sensor. Biosensors 2:71–87
Dill KA (1997) Additivity principles in biochemistry. J Biol Chem 272:701–704
Dokter P, Frank J, Duine JA (1986) Purification and characterization of quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus L.M.D. 79.41. Biochem J 239:163–167
Fang TY, Honzatko RB, Reilly PJ, Ford C (1998) Mutations to alter Aspergillus awamori glucoamylase selectivity. II. Mutation of residues 119 and 121. Protein Eng 11:127–133
Hamamatsu N, Aita T, Nomiya Y, Uchiyama H, Nakajima M, Husimi Y, Shibanaka Y (2005) Biased mutation-assembling: an efficient method for rapid directed evolution through simultaneous mutation accumulation. Protein Eng Des Sel 18:265–271
Hauge JG (1964) Glucose dehydrogenase of Bacterium anitratum: an enzyme with a novel prosthetic group. J Biol Chem 239:3630–3639
Hogrefe HH, Cline J, Youngblood GL, Allen RM (2002) Creating randomized amino acid libraries with the QuikChange Multi Site-Directed Mutagenesis Kit. Biotechniques 33:1158–1160
Igarashi S, Hirokawa T, Sode K (2004a) Engineering PQQ glucose dehydrogenase with improved substrate specificity: site-directed mutagenesis studies on the active center of PQQ glucose dehydrogenase. Biomol Eng 21:81–89
Igarashi S, Okuda J, Ikebukuro K, Sode K (2004b) Molecular engineering of PQQGDH and its applications. Arch Biochem Biophys 428:52–63
Kegler-Ebo DM, Docktor CM, DiMaio D (1994) Codon cassette mutagenesis: a general method to insert or replace individual codons by using universal mutagenic cassettes. Nucleic Acids Res 22:1593–1599
Klibanov AM (1990) Asymmetric transformations catalyzed by enzymes in organic-solvents. Acc Chem Res 23:114–120
Leung DW, Chen E, Goeddel DV (1989) A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11–15
Ye L, Haemmerle M, Olsthoorn AJJ, Schuhmann W, Schmidt HL, Duine JA, Heller A (1993) High current density “wired” quinoprotein glucose dehydrogenase electrode. Anal Chem 65:238–241
Murakami H, Hohsaka T, Sisido M (2002) Random insertion and deletion of arbitrary number of bases for codon-based random mutation of DNAs. Nat Biotechnol 20:76–81
Olsthoorn AJ, Otsuki T, Duine JA (1998) Negative cooperativity in the steady-state kinetics of sugar oxidation by soluble quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus. Eur J Biochem 255:255–261
Oubrie A, Rozeboom HJ, Kalk KH, Olsthoorn AJJ, Duine JA, Dijkstra BW (1999) Structure and mechanism of soluble quinoprotein glucose dehydrogenase. EMBO J 18:5187–5194
Reddy SY, Bruice TC (2004) Mechanism of glucose oxidation by quinoprotein soluble glucose dehydrogenase: insights from molecular dynamics studies. J Am Chem Soc 126:2431–2438
Uchiyama H, Inaoka T, Ohkuma-Soyejima T, Togame H, Shibanaka Y, Yoshimoto T, Kokubo T (2000) Directed evolution to improve the thermostability of prolyl endopeptidase. J Biochem 128:441–447
Wells JA (1990) Additivity of mutational effects in proteins. Biochemistry 29:8509–8517
Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115
Acknowledgement
This work was performed as a part of the R&D Project of the Industrial Science and Technology Frontier Program supported by New Energy and Industrial Technology Development Organization in Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hamamatsu, N., Suzumura, A., Nomiya, Y. et al. Modified substrate specificity of pyrroloquinoline quinone glucose dehydrogenase by biased mutation assembling with optimized amino acid substitution. Appl Microbiol Biotechnol 73, 607–617 (2006). https://doi.org/10.1007/s00253-006-0521-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0521-4