Abstract
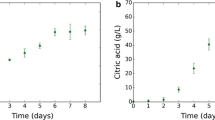
It is well-known that secondary metabolite production is repressed by excess nitrogen substrate available in the fermentation media. Although the nitrogen catabolite repression has been known, quantitative process models have not been reported to represent this phenomenon in complex medium. In this paper, we present a cybernetic model for rifamycin B production via Amycolatopsis mediterranei S699 in complex medium, which is typically used in industry. Nitrogen substrate is assumed to be present in two forms in the medium; available nitrogen (S ANS) such as free amino acids and unavailable nitrogen (S UNS) such as peptides and proteins. The model assumes that an inducible enzyme catalyzes the conversion of S UNS to S ANS. Although S ANS is required for growth and product formation, high concentrations were found to inhibit rifamycin production. To experimentally validate the model, five different organic nitrogen sources were used that differ in the ratio of S ANS/S UNS. The model successfully predicts higher rifamycin B productivity for nitrogen sources that contain lower initial S ANS. The higher productivity is attributed to the sustained availability of S ANS at low concentration via conversion of S UNS to S ANS, thereby minimizing the effects of nitrogen catabolite repression on rifamycin production. The model can have applications in model-based optimization of substrate feeding recipe and in monitoring and control of fed batch processes.



Similar content being viewed by others
References
Aharanowitz Y (1980) Nitrogen metabolite regulation of antibiotic biosynthesis. Annu Rev Microbiol 34:9–33
Aharanowitz Y, Demain AL (1979) Nitrogen nutrition and regulation of cephalosporin production in Streptomyces clavuligerus. Can J Microbiol 25:61–67
Bajapai RK, Reuß M (1981) Evaluation of feeding strategies in carbon regulated secondary metabolite production through mathematical modeling. Biotechnol Bioeng 23:739–763
Bapat PM, Wangikar PP (2004) Optimization of Rifamycin B fermentation in shake flasks via a machine-learning-based approach. Biotechnol Bioeng 86:201–208
Bapat PM, Kundu S, Wangikar PP (2003) An optimized method for Aspergillus niger spore production on natural carrier substrates. Biotech Prog 19:1683–1688
Bapat PM, Nandy S, Wangikar P, Venkatesh KV (2005) Quantification of metabolically active biomass using Methylene Blue dye Reduction Test (MBRT): measurement of CFU in about 200 s. J Microbiol Methods DOI 10.1016/j.mimet.2005.06.010
Bapat PM, Bhartiya S, Venkatesh KV, Wangikar PP (2006) A structured kinetic model to represent the utilization of multiple substrates in complex media during rifamycin B fermentation. Biotechnol Bioeng DOI 10.1002/bit.20767
Bascaran V, Hardisson C, Brana A (1989) Regulation of nitrogen catebolite enzymes in Streptomyces clavuligerus. J Gen Microbiol 135:2465–2474
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72:248–254
Brana AF, Demain AL (1988) Nitrogen control of antibiotic biosynthesis in actinomycetes. In: Sanchez SE, Boca RFL (eds) Nitrogen source control of microbial processes. CRC, London, UK, pp 99–119
Calam CT (1987) Process development in antibiotic fermentations. University of Cambridge, Cambridge, pp 1–217
Dhurjati P, Ramkrishna D, Flickinger MC, Tsao GT (1985) A cybernetic view of microbial growth: modeling of cells as optimal strategists. Biotechnol Bioeng 27:1–9
Dynesen J, Smits HP, Olsson L, Nielsen J (1998) Carbon catabolite repression of invertase during batch cultivations of Saccharomyces cerevisiae: the role of glucose, fructose, and mannose. Appl Microbiol Biotechnol 50:579–582
El-Tayeb OM, Salama AA, Hussein MMM, El-Sedawy HF (2004) Optimization of industrial production of rifamycin B by Amycolatopsis mediterranei. I. The role of colony morphology and nitrogen sources in productivity. Afr J Biotechnol 3:266–272
Kawahuchi T, Asahi T, Satoh T, Uozumi T, Beppu T (1984) B factor, an essential regulatory substance inducing the production of rifamycin in Nocardia sp. J Antibiotics 37:1587–1595
Kim CG, Kirschning A, Bergon P, Zhou P, Su E, Sauerbrei B, Ning S, Ahn Y, Breuer M, Leistner E (1996) Biosynthesis of 3-amino-5-hydroxybenzoic acid, the precursor of mC(7)N units in ansamycin antibiotics. JACS 118:7486–7491
Kompala DS, Ramakrishna D, Tsao GT (1984) Cybernetic modelling of microbial growth in simple substrates. Biotechnol Bioeng 26:1272–1281
Krabben P, Theodorus D (2001) Fermentation process to produce clavulanic acid at a low concentration of free amino acids. WO Patent 00/01840
Martin JF, Demain AL (1980) Control of antibiotic synthesis. Microbiol Rev 44:230–251
Moore S (1968) Amino acid analysis: aqueous dimethyl sulfoxide as solvent for the ninhydrin reaction. J Biol Chem 243:6281–6283
Nielsen J (1995) Physiological engineering aspect of Penicillium chrysogenum. Polyteknisk Forlag, Lingby, Denmark
Pasqualucci CR, Radaelli P, Gallo GG (1970) Improved differential spectrophotometric determination of rifamycins. J Pharm Sci 59:685–687
Patnaik PR (2001) Penicillin fermentation: mechanisms and models for industrial-scale bioreactors. Crit Rev Microbiol 27:25–39
Ramakrishna R, Ramkrishna D, Konopka AE (1997) Microbial growth on substitutable substrates: characterizing the consumer–resource relationship. Biotechnol Bioeng 54:77–90
Ross LF, Chapital DC (1987) Simultaneous determination of carbohydrates and products of carbohydrate metabolism in fermentation mixtures by HPLC. J Chromatogr Sci 25:112–117
Sanchez S, Demain AL (2002) Metabolic regulation of fermentation process. Enzyme Microbiol Technol 31:895–906
Sensi P, Thiemann JE (1967) Production of rifamycins. Prog Ind Microbiol 6:21–59
Sepkowitz KA, Rafalli J, Riley L, Kiehn TE, Armstrong D (1995) Tuberculosis in the AIDS era. Clin Microbiol Rev 8:180-199
Stanbury PF, Whitaker A (1984) The development of inocula for industrial fermentations. In: Principles of fermentation technology. Oxford Pergamon, pp 108–119
Stratmann A, Schupp T, Toupet C, Schilling W, Oberer L, Traber R (2002) New insights into rifamycin B biosynthesis: isolation of proansamycin B and 34a-deoxy-rifamycin W as early macrocyclic intermediates indicating two separated biosynthetic pathways. J Antibiotics 55:396–406
Untrau S, Lebrihi A, Lefebvre G, Germain P (1994) Nitrogen catebolite regulation of Spiramycin production in Streptomyces ambofaciens. Curr Microbiol 28:111–118
Venkateswarlu G, Murali Krishna PS, Rao VL (1999) Production of rifamycin using Amycolatopsis mediterranei (MTCC14). Bioprocess Eng 20:27–30
Vining LC, Doull JL (1988) Catebolite repression of secondary metabolism in actinomycetes. In: Okami M (eds) 7th International symposium on Biology of Actinomycetes. Japan Scientific Societies, Tokyo, pp 406–411
Yu TW, Muller R, Muller M, Zhang XH, Draeger G, Kim CG, Leistner E, Floss HG (2001) Mutational analysis and reconstituted expression of the biosynthetic genes involved in the formation of 3-amino-5-hydroxybenzoic acid, the starter unit of rifamycin biosynthesis in Amycolatopsis mediterranei S699. J Biol Chem 276:12546–12555
Zimbro MJ, Power DA (2003) (eds) Difco and BBL manual of microbiological culture media. Becton, Dickinson and company, Maryland, USA, pp 686–687
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bapat, P.M., Sohoni, S.V., Moses, T.A. et al. A cybernetic model to predict the effect of freely available nitrogen substrate on rifamycin B production in complex media. Appl Microbiol Biotechnol 72, 662–670 (2006). https://doi.org/10.1007/s00253-006-0341-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-006-0341-6