Abstract
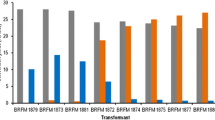
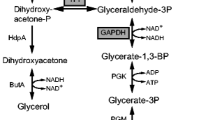
Rhizopus oryzae is capable of producing high levels of lactic acid by the fermentation of glucose. Yields typically vary over 60–80%, with the remaining glucose diverted primarily into ethanol fermentation. The goal of this work was to increase lactate dehydrogenase (LDH) activity, so lactic acid fermentation could more effectively compete for available pyruvate. Three different constructs, pLdhA71X, pLdhA48XI, and pLdhA89VII, containing various lengths of the ldhA gene fragment, were transformed into R. oryzae. This fungus rarely integrates DNA used for transformation, but instead relies on extra-chromosomal replication in a high-copy number. Plasmid pLdhA48XI was linearized prior to transformation in order to facilitate integration into the pyrG gene used for selection. Isolates transformed with ldhA containing plasmid were compared with both the wild-type parent strain and the auxotrophic recipient strain containing vector only. All isolates transformed with pLdhA71X or pLdhA48XI had multiple copies of the ldhA gene that resulted in ldhA transcript accumulation, LDH specific activity, and lactic acid production higher than the controls. Integration of plasmid pLdhA48XI increased the stability of the strain, but did not seem to offer any benefit for increasing lactic acid production. Since lactic acid fermentation competes with ethanol and fumaric acid production, it was not unexpected that increased lactic acid production was always concomitant with decreased ethanol and fumaric acid. Plasmid pLdhA71X, containing a large ldhA fragment (6.1 kb), routinely yielded higher levels of lactic acid than the smaller region (3.3 kb) used to construct plasmid pLdhA48XI. The greatest levels of ldhA transcript and enzyme production occurred with isolates transformed with plasmid pLdhA89VII. However, these transformants always produced less lactic acid and higher amounts of ethanol, fumaric, and glycerol compared with the control.




Similar content being viewed by others
References
Adachi E, Torigoe M, Sugiyama M, Nikawa JI, Shimizu K (1988) Modification of metabolic pathways of Saccharomyces cerevisiae by expression of lactate dehydrogenase and deletion of pyruvate decarboxylase genes for the lactic acid fermentation at low pH. J Ferment Bioeng 86:284–289
Bakker BM, Overkamp KM, Maris AJA van, Kotter P, Luttik MA H, Dijken JP van, Pronk JT (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25:15–37
Bunch PK, Mat-Jan F, Lee N, Clark DP (1997) The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187–195
Dequin S, Barre P (1994) Mixed lactic acid-alcoholic fermentation by Saccharomyces cerevisiae expressing the Lactobacillus casei l(+)-LDH. Biotechnology 12:173–177
Friedberg D, Peleg Y, Monsonego A, Maissi S, Battat E, Rokem JS, Goldberg I (1995) The fumR gene encoding fumarase in the filamentous fungus Rhizopus oryzae: cloning, structure, and expression. Gene 163:139–144
Hester A (2000) IB Market Forecast. Ind Bioprocess 22:3–5
Kenealy W, Zaady E, DuPreez JC, Stieglitz B, Goldberg I (1986) Biochemical aspects of fumaric acid accumulation by Rhizopus arrhizus. Appl Environ Microbiol 52:128–133
Osmani SA, Scrutton MC (1985) The sub-cellular localisation and regulatory properties of pyruvate carboxylase from Rhizopus arrhizus. Eur J Biochem 147:119–128
Peleg Y, Battat E, Scrutton MC, Goldberg I (1989) Isozyme pattern and subcellular localisation of enzyme involved in fumaric acid accumulation by Rhizopus oryzae. Appl Microbiol Biotechnol 32:334–339
Porro D, Bianchi MM, Brambilla L, Menghini R, Bolzani D, Carrera V, Lievense J, Liu CL, Ranzi BM, Frontali L, Alberghina L (1999) Replacement of a metabolic pathway for large-scale production of lactic acid from engineered yeasts. Appl Environ Microbiol 65:4211–4215
Skory CD (2000) Isolation and expression of lactate dehydrogenase genes from Rhizopus oryzae. Appl Environ Microbiol 66:2343–2348
Skory CD (2001) Fungal lactate dehydrogenase gene and constructs for the expression thereof. US Patent 6,268,189
Skory CD (2002) Homologous recombination and double-strand break repair in the transformation of Rhizopus oryzae. Mol Genet Genomics 268:397–406
Skory CD (2003a) Induction of Rhizopus oryzae pyruvate decarboxylase genes. Curr Microbiol 47:59–64
Skory CD (2003b) Lactic acid production by Saccharomyces cerevisiae expressing a Rhizopus oryzae lactate dehydrogenase gene. J Ind Microbiol Biotechnol 30:22–27
Taherzadeh MJ, Adler L, Liden G (2002) Strategies for enhancing fermentative production of glycerol—a review. Enzyme Microb Technol 31:53–66
Watkins K (2002) A solvent business. Chem Eng News 80:15–16
Author information
Authors and Affiliations
Corresponding author
Additional information
Disclaimer:
Names are necessary to report factually on available data; however, the USDA neither guarantees nor warrants the standard of the product, and the use of a name by the USDA implies no approval of the product to the exclusion of others that may also be suitable.
Rights and permissions
About this article
Cite this article
Skory, C.D. Lactic acid production by Rhizopus oryzae transformants with modified lactate dehydrogenase activity. Appl Microbiol Biotechnol 64, 237–242 (2004). https://doi.org/10.1007/s00253-003-1480-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-003-1480-7