Abstract
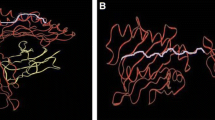
HLA-DQ molecules have been associated with susceptibility to a number of autoimmune and other diseases, possibly through the peptide repertoire that can be presented by different allelic products. It is thus of importance to understand which peptides can be bound by different HLA-DQ allelic products. Recently, a model for HLA-DQ has been described and used to derive peptide positional environments for HLA-DQ allelic products. By combining the peptide positional environments with known HLA-DQ peptide binding motifs, a set of predictions of likely anchor motifs for many of the products of HLA-DQ allelic variants are made and presented in a table referred to as a roadmap for HLA-DQ peptide binding specificities.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 3 March 1999 / Revised: 20 May 1999
Rights and permissions
About this article
Cite this article
Baas, A., Gao, X. & Chelvanayagam, G. Peptide binding motifs and specificities for HLA-DQ molecules. Immunogenetics 50, 8–15 (1999). https://doi.org/10.1007/s002510050680
Issue Date:
DOI: https://doi.org/10.1007/s002510050680