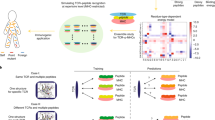
Abstract
Recent advances in molecular and bioinformatic methods have greatly improved our ability to study the formation of an adaptive immune response towards foreign pathogens, self-antigens, and cancer neoantigens. T cell receptors (TCR) are the key players in this process that recognize peptides presented by major histocompatibility complex (MHC). Owing to the huge diversity of both TCR sequence variants and peptides they recognize, accumulation and complex analysis of large amounts of TCR-antigen specificity data is required for understanding the structure and features of adaptive immune responses towards pathogens, vaccines, cancer, as well as autoimmune responses. In the present review, we summarize recent efforts on gathering and interpreting TCR-antigen specificity data and outline the critical role of tighter integration with other immunoinformatics data sources that include epitope MHC restriction, TCR repertoire structure models, and TCR/peptide/MHC structural data. We suggest that such integration can lead to the ability to accurately annotate individual TCR repertoires, efficiently estimate epitope and neoantigen immunogenicity, and ultimately, in silico identify TCRs specific to yet unstudied antigens and predict self-peptides related to autoimmunity.

Similar content being viewed by others
References
Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML (2010) Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 115:3718–3725. https://doi.org/10.1182/blood-2009-10-251124
Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams M, Bell JI, McMichael A, Davis MM (1996) Phenotypic analysis of antigen-specific T lymphocytes. Science 274:94–96. https://doi.org/10.1126/science.274.5284.94
Bacher P, Scheffold A (2013) Flow-cytometric analysis of rare antigen-specific T cells. Cytometry A 83A:692–701. https://doi.org/10.1002/cyto.a.22317
Bagaev DV, Zvyagin IV, Putintseva EV, Izraelson M, Britanova OV, Chudakov DM, Shugay M (2016) VDJviz: a versatile browser for immunogenomics data. BMC Genomics 17(1):453. https://doi.org/10.1186/s12864-016-2799-7
Bagaev DV, Vroomans RMA, Samir J, Stervbo U, Rius C, Dolton G, Greenshields-Watson A, Attaf M, Egorov ES, Zvyagin IV, Babel N, Cole DK, Godkin AJ, Sewell AK, Kesmir C, Chudakov DM, Luciani F, Shugay M (2019) VDJdb in 2019: database extension, new analysis infrastructure and a T-cell receptor motif compendium. Nucleic Acids Res. https://doi.org/10.1093/nar/gkz874
Benichou J, Ben-Hamo R, Louzoun Y, Efroni S (2012) Rep-Seq: uncovering the immunological repertoire through next-generation sequencing. Immunology 135(3):183–191. https://doi.org/10.1111/j.1365-2567.2011.03527.x
Bentzen AK, Marquard AM, Lyngaa R, Saini SK, Ramskov S, Donia M, Such L, Furness AJ, McGranahan N, Rosenthal R, Straten PT, Szallasi Z, Svane IM, Swanton C, Quezada SA, Jakobsen SN, Eklund AC, Hadrup SR (2016) Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat Biotechnol 34:1037–1045. https://doi.org/10.1038/nbt.3662
Birnbaum ME, Mendoza JL, Sethi DK, Dong S, Glanville J, Dobbins J, Ozkan E, Davis MM, Wucherpfennig KW, Garcia KC (2014) Deconstructing the peptide-MHC specificity of T cell recognition. Cell 157:1073–1087. https://doi.org/10.1016/j.cell.2014.03.047
Bjerregaard AM, Nielsen M, Hadrup SR, Szallasi Z, Eklund AC (2017) MuPeXI: prediction of neo-epitopes from tumor sequencing data. Cancer Immunol Immunother 66(9):1123–1130. https://doi.org/10.1007/s00262-017-2001-3
Borrman T, Cimons J, Cosiano M, Purcaro M, Pierce BG, Baker BM, Weng Z (2017) ATLAS: A database linking binding affinities with structures for wild-type and mutant TCR-pMHC complexes. Proteins 85:908–916. https://doi.org/10.1002/prot.25260
Britanova OV, Shugay M, Merzlyak EM et al (2016) Dynamics of Individual T Cell Repertoires: From Cord Blood to Centenarians. J Immunol 196:5005–5013. https://doi.org/10.4049/jimmunol.1600005
Calis JJA, Maybeno M, Greenbaum JA et al (2013) Properties of MHC Class I Presented Peptides That Enhance Immunogenicity. PLOS Comput Biol 9:e1003266. https://doi.org/10.1371/journal.pcbi.1003266
Chen X, Poncette L, Blankenstein T (2017) Human TCR-MHC coevolution after divergence from mice includes increased nontemplate-encoded CDR3 diversity. J Exp Med 214:3417–3433. https://doi.org/10.1084/jem.20161784
Chowell D, Krishna S, Becker PD, Cocita C, Shu J, Tan X, Greenberg PD, Klavinskis LS, Blattman JN, Anderson KS (2015) TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc Natl Acad Sci 112:E1754–E1762. https://doi.org/10.1073/pnas.1500973112
Dash P, Fiore-Gartland AJ, Hertz T et al (2017) Quantifiable predictive features define epitope specific T cell receptor repertoires. Nature 547:89–93. https://doi.org/10.1038/nature22383
DeWitt WS, Smith A, Schoch G et al (2018) Human T cell receptor occurrence patterns encode immune history, genetic background, and receptor specificity. eLife 7. https://doi.org/10.7554/eLife.38358
Dolton G, Tungatt K, Lloyd A et al (2015) More tricks with tetramers: a practical guide to staining T cells with peptide–MHC multimers. Immunology 146:11–22. https://doi.org/10.1111/imm.12499
Egorov ES, Kasatskaya SA, Zubov VN et al (2018) The Changing Landscape of Naive T Cell Receptor Repertoire With Human Aging. Front Immunol 9. https://doi.org/10.3389/fimmu.2018.01618
Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, Desmarais C, Klinger M, Carlson CS, Hansen JA, Rieder M, Robins HS (2017) Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet 49:659–665. https://doi.org/10.1038/ng.3822
Gee MH, Han A, Lofgren SM et al (2018) Antigen identification for orphan T cell receptors expressed on tumor-infiltrating lymphocytes. Cell 172:549–563. https://doi.org/10.1016/j.cell.2017.11.043
Gielis S, Moris P, Neuter ND et al (2018) TCRex: a webtool for the prediction of T-cell receptor sequence epitope specificity. bioRxiv:373472. https://doi.org/10.1101/373472
Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, Haas N, Arlehamn CSL, Sette A, Boyd SD, Scriba TJ, Martinez OM, Davis MM (2017) Identifying specificity groups in the T cell receptor repertoire. Nature 547:94–98. https://doi.org/10.1038/nature22976
Gowthaman R, Pierce BG (2018) TCRmodel: high resolution modeling of T cell receptors from sequence. Nucleic Acids Res 46:W396–W401. https://doi.org/10.1093/nar/gky432
Gowthaman R, Pierce BG (2019) TCR3d: The T cell receptor structural repertoire database. Bioinforma Oxf Engl. https://doi.org/10.1093/bioinformatics/btz517
Hoffmann T, Marion A, Antes I (2017) DynaDom: structure-based prediction of T cell receptor inter-domain and T cell receptor-peptide-MHC (class I) association angles. BMC Struct Biol 17:2. https://doi.org/10.1186/s12900-016-0071-7
Jokinen E, Huuhtanen J, Mustjoki S et al (2019) Determining epitope specificity of T cell receptors with TCRGP. bioRxiv:542332. https://doi.org/10.1101/542332
Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M (2017) NetMHCpan-4.0: Improved Peptide-MHC Class I Interaction Predictions Integrating Eluted Ligand and Peptide Binding Affinity Data. J Immunol 199:3360–3368. https://doi.org/10.4049/jimmunol.1700893
Jurtz VI, Jessen LE, Bentzen AK et al (2018) NetTCR: sequence-based prediction of TCR binding to peptide-MHC complexes using convolutional neural networks. bioRxiv:433706. https://doi.org/10.1101/433706
Kar P, Ruiz-Perez L, Arooj M, Mancera RL (2018) Current methods for the prediction of T-cell epitopes. Pept Sci 110:e24046. https://doi.org/10.1002/pep2.24046
Klinger M, Pepin F, Wilkins J, Asbury T, Wittkop T, Zheng J, Moorhead M, Faham M (2015) Multiplex Identification of Antigen-Specific T Cell Receptors Using a Combination of Immune Assays and Immune Receptor Sequencing. PLoS One. 10:e0141561. https://doi.org/10.1371/journal.pone.0141561 eCollection 2015
Komech EA, Pogorelyy MV, Egorov ES et al (2018) CD8+ T cells with characteristic T cell receptor beta motif are detected in blood and expanded in synovial fluid of ankylosing spondylitis patients. Rheumatology 57:1097–1104. https://doi.org/10.1093/rheumatology/kex517
Konstantinou GN (2017) T-Cell Epitope Prediction. Methods Mol Biol Clifton NJ 1592:211–222. https://doi.org/10.1007/978-1-4939-6925-8_17
Kula T, Dezfulian MH, Wang CI et al (2019) T-Scan: a genome-wide method for the systematic discovery of T cell epitopes. Cell 178:1016–1028. https://doi.org/10.1016/j.cell.2019.07.009
Leem J, de Oliveira SHP, Krawczyk K, Deane CM (2018) STCRDab: the structural T-cell receptor database. Nucleic Acids Res 46:D406–D412. https://doi.org/10.1093/nar/gkx971
Mahajan S, Vita R, Shackelford D et al (2018) Epitope Specific Antibodies and T Cell Receptors in the Immune Epitope Database. Front Immunol 9. https://doi.org/10.3389/fimmu.2018.02688
Marcou Q, Mora T, Walczak AM (2018) High-throughput immune repertoire analysis with IGoR. Nat Commun 9:561. https://doi.org/10.1038/s41467-018-02832-w
Mora T, Walczak AM (2016) Quantifying lymphocyte receptor diversity. bioRxiv:046870. https://doi.org/10.1101/046870
Moritz A, Anjanappa R, Wagner C et al (2019) High-throughput peptide-MHC complex generation and kinetic screenings of TCRs with peptide-receptive HLA-A*02:01 molecules. Sci Immunol 4:eaav0860. https://doi.org/10.1126/sciimmunol.aav0860
Murugan A, Mora T, Walczak AM, Callan CG (2012) Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci 109:16161–16166. https://doi.org/10.1073/pnas.1212755109
Napolitani G, Kurupati P, Teng KWW, Gibani MM, Rei M, Aulicino A, Preciado-Llanes L, Wong MT, Becht E, Howson L, de Haas P, Salio M, Blohmke CJ, Olsen LR, Pinto DMS, Scifo L, Jones C, Dobinson H, Campbell D, Juel HB, Thomaides-Brears H, Pickard D, Bumann D, Baker S, Dougan G, Simmons A, Gordon MA, Newell EW, Pollard AJ, Cerundolo V (2018) Clonal analysis of Salmonella-specific effector T cells reveals serovar-specific and cross-reactive T cell responses. Nat Immunol 19(7):742–754. https://doi.org/10.1038/s41590-018-0133-z
Neller MA, Ladell K, McLaren JE, Matthews KK, Gostick E, Pentier JM, Dolton G, Schauenburg AJ, Koning D, Fontaine Costa AI, Watkins TS, Venturi V, Smith C, Khanna R, Miners K, Clement M, Wooldridge L, Cole DK, van Baarle D, Sewell AK, Burrows SR, Price DA, Miles JJ (2015) Naive CD8+ T-cell precursors display structured TCR repertoires and composite antigen-driven selection dynamics. Immunol Cell Biol 93:625–633. https://doi.org/10.1038/icb.2015.17
Ng AHC, Peng S, Xu AM, Noh WJ, Guo K, Bethune MT, Chour W, Choi J, Yang S, Baltimore D, Heath JR (2019) MATE-Seq: microfluidic antigen-TCR engagement sequencing. Lab Chip. 19:3011–3021. https://doi.org/10.1039/c9lc00538b
Pogorelyy MV, Shugay M (2019) A framework for annotation of antigen specificities in high-throughput T-cell repertoire sequencing studies. bioRxiv:676239. https://doi.org/10.1101/676239
Pogorelyy MV, Fedorova AD, McLaren JE, Ladell K, Bagaev DV, Eliseev AV, Mikelov AI, Koneva AE, Zvyagin IV, Price DA, Chudakov DM, Shugay M (2018) Exploring the pre-immune landscape of antigen-specific T cells. Genome Med 10:68. https://doi.org/10.1186/s13073-018-0577-7
Pogorelyy MV, Minervina AA, Shugay M, Chudakov DM, Lebedev YB, Mora T, Walczak AM (2019) Detecting T cell receptors involved in immune responses from single repertoire snapshots. PLOS Biol 17:e3000314. https://doi.org/10.1371/journal.pbio.3000314
Ritvo P-G, Saadawi A, Barennes P, Quiniou V, Chaara W, el Soufi K, Bonnet B, Six A, Shugay M, Mariotti-Ferrandiz E, Klatzmann D (2018) High-resolution repertoire analysis reveals a major bystander activation of Tfh and Tfr cells. Proc Natl Acad Sci 115:9604–9609. https://doi.org/10.1073/pnas.1808594115
Rius C, Attaf M, Tungatt K et al (2018) Peptide–MHC Class I Tetramers Can Fail To Detect Relevant Functional T Cell Clonotypes and Underestimate Antigen-Reactive T Cell Populations. J Immunol. https://doi.org/10.4049/jimmunol.1700242
Rossjohn J, Gras S, Miles JJ et al (2015) T Cell Antigen Receptor Recognition of Antigen-Presenting Molecules. Annu Rev Immunol 33:169–200. https://doi.org/10.1146/annurev-immunol-032414-112334
Rubelt F, Busse CE, Bukhari SAC et al (2017) Adaptive Immune Receptor Repertoire Community recommendations for sharing immune-repertoire sequencing data. Nat Immunol 18(12):1274–1278. https://doi.org/10.1038/ni.3873
Rubinsteyn A, Hodes I, Kodysh J et al (2018) Vaxrank: A computational tool for designing personalized cancer vaccines. bioRxiv:142919. https://doi.org/10.1101/142919
Saade F, Gorski SA, Petrovsky N (2012) Pushing the frontiers of T-cell vaccines: accurate measurement of human T-cell responses. Expert Rev Vaccines 11:1459–1470. https://doi.org/10.1586/erv.12.125
Saini SK, Tamhane T, Anjanappa R et al (2019) Empty peptide-receptive MHC class I molecules for efficient detection of antigen-specific T cells. Sci Immunol 4:eaau9039. https://doi.org/10.1126/sciimmunol.aau9039
Schenck RO, Lakatos E, Gatenbee C, Graham TA, Anderson ARA (2019) NeoPredPipe: high-throughput neoantigen prediction and recognition potential pipeline. BMC Bioinformatics 20:264. https://doi.org/10.1186/s12859-019-2876-4
Schritt D, Li S, Rozewicki J et al (2019) Repertoire Builder: high-throughput structural modeling of B and T cell receptors. Mol Syst Des Eng 4:761–768. https://doi.org/10.1039/C9ME00020H
Sethna Z, Elhanati Y, Callan CG et al (2019) OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs. Bioinforma Oxf Engl. https://doi.org/10.1093/bioinformatics/btz035
Sharma G, Rive CM, Holt RA (2019) Rapid selection and identification of functional CD8+ T cell epitopes from large peptide-coding libraries. Nat Commun 10:4553. https://doi.org/10.1038/s41467-019-12444-7
Sharon E, Sibener LV, Battle A, Fraser HB, Garcia KC, Pritchard JK (2016) Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat Genet 48:995–1002. https://doi.org/10.1038/ng.3625
Sharrock CE, Kaminski E, Man S (1990) Limiting dilution analysis of human T cells: a useful clinical tool. Immunol Today 11:281–286
Shugay M, Bolotin DA, Putintseva EV, Pogorelyy MV, Mamedov IZ, Chudakov DM (2013) Huge overlap of individual TCR beta repertoires. Front Immunol 4:466. https://doi.org/10.3389/fimmu.2013.00466
Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, Pogorelyy MV, Nazarov VI, Zvyagin IV, Kirgizova VI, Kirgizov KI, Skorobogatova EV, Chudakov DM (2015) VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLOS Comput Biol 11:e1004503. https://doi.org/10.1371/journal.pcbi.1004503
Shugay M, Bagaev DV, Zvyagin IV, Vroomans RM, Crawford JC, Dolton G, Komech EA, Sycheva AL, Koneva AE, Egorov ES, Eliseev AV, van Dyk E, Dash P, Attaf M, Rius C, Ladell K, McLaren J, Matthews KK, Clemens EB, Douek DC, Luciani F, van Baarle D, Kedzierska K, Kesmir C, Thomas PG, Price DA, Sewell AK, Chudakov DM (2018) VDJdb: a curated database of T-cell receptor sequences with known antigen specificity. Nucleic Acids Res 46:D419–D427. https://doi.org/10.1093/nar/gkx760
Siewert K, Malotka J, Kawakami N, Wekerle H, Hohlfeld R, Dornmair K (2011) Unbiased identification of target antigens of CD8+ T cells with combinatorial libraries coding for short peptides. Nat Med 18:824–828. https://doi.org/10.1038/nm.2720
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, Cancer Genome Atlas Research Network, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I (2018) The Immune Landscape of Cancer. Immunity 48:812–830. https://doi.org/10.1016/j.immuni.2018.03.023
Tickotsky N, Sagiv T, Prilusky J et al (2017) McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinforma Oxf Engl 33:2924–2929. https://doi.org/10.1093/bioinformatics/btx286
Venturi V, Price DA, Douek DC, Davenport MP (2008) The molecular basis for public T-cell responses? Nat Rev Immunol 8:231–238. https://doi.org/10.1038/nri2260
Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, Peters B (2015) The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43:D405–D412. https://doi.org/10.1093/nar/gku938
Zhang SQ, Ma KY, Schonnesen AA et al (2018) High-throughput determination of the antigen specificities of T cell receptors in single cells. Nat Biotechnol 36:1156–1159. https://doi.org/10.1038/nbt.4282
Zvyagin IV, Pogorelyy MV, Ivanova ME et al (2014) Distinctive properties of identical twins' TCR repertoires revealed by high-throughput sequencing. Proc Natl Acad Sci U S A 111(16):5980–5985. https://doi.org/10.1073/pnas.1319389111
Acknowledgments
We would like to thank anonymous reviewers for their valuable suggestions and comments.
Funding
This work was supported by RFBR grant No 19-34-70011.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on “Nomenclature, databases and bioinformatics in Immunogenetics”
Rights and permissions
About this article
Cite this article
Zvyagin, I.V., Tsvetkov, V.O., Chudakov, D.M. et al. An overview of immunoinformatics approaches and databases linking T cell receptor repertoires to their antigen specificity. Immunogenetics 72, 77–84 (2020). https://doi.org/10.1007/s00251-019-01139-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-019-01139-4