Abstract
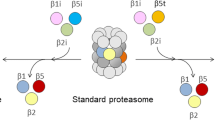
The thymoproteasome is a recently discovered, specialized form of 20S proteasomes expressed exclusively in the thymic cortex. Although the precise molecular mechanism by which the thymoproteasome exerts its function remains to be elucidated, accumulating evidence indicates that it plays a crucial role in positive selection of T cells. In the present study, we analyzed the evolution of the β5t subunit, a β-type catalytic subunit uniquely present in thymoproteasomes. The gene coding for the β5t subunit, designated PSMB11, was identified in the cartilaginous fish, the most divergent group of jawed vertebrates compared to the other jawed vertebrates, but not in jawless vertebrates or invertebrates. Interestingly, teleost fish have two copies of apparently functional PSMB11 genes, designated PSMB11a and PSMB11b, that encode β5t subunits with distinct amino acids in the S1 pocket. BLAST searches of genome databases suggest that birds such as chickens, turkey, and zebra finch lost the PSMB11 gene, and have neither thymoproteasomes nor immunoproteasomes. In mammals, reptiles, amphibians, and teleost fishes, the PSMB11 gene (the PSMB11a gene in teleost fish) is located next to the PSMB5 gene coding for the β5 subunit of the standard 20S proteasome, indicating that the PSMB11 gene arose by tandem duplication from the evolutionarily more ancient PSMB5 gene. The general absence of introns in PSMB11 and an unusual exon–intron structure of jawed vertebrate PSMB5 suggest that PSMB5 lost introns and duplicated in tandem in a common ancestor of jawed vertebrates, with PSMB5 subsequently gaining two introns and PSMB11 remaining intronless.





Similar content being viewed by others
References
Abdulla S, Beck S, Belich M, Jackson A, Nakamura T, Trowsdale J (1996) Divergent intron arrangement in the MB1/LMP7 proteasome gene pair. Immunogenetics 44:254–258
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195–201
Bajoghli B, Guo P, Aghaallaei N, Hirano M, Strohmeier C, McCurley N, Bockman DE, Schorpp M, Cooper MD, Boehm T (2011) A thymus candidate in lampreys. Nature 470:90–94
Balakrishnan CN, Ekblom R, Volker M, Westerdahl H, Godinez R, Kotkiewicz H, Burt DW, Graves T, Griffin DK, Warren WC, Edwards SV (2010) Gene duplication and fragmentation in the zebra finch major histocompatibility complex. BMC Biol 8:29
Belich MP, Trowsdale J (1995) Proteasome and class I antigen processing and presentation. Mol Biol Rep 21:53–56
Chaves LD, Krueth SB, Reed KM (2009) Defining the turkey MHC: sequence and genes of the B locus. J Immunol 183:6530–6537
Clark MS, Pontarotti P, Gilles A, Kelly A, Elgar G (2000) Identification and characterization of a β proteasome subunit cluster in the Japanese pufferfish (Fugu rubripes). J Immunol 165:4446–4452
Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Le Blomberg A, Bouffard P, Burt DW, Crasta O, Crooijmans RP, Cooper K, Coulombe RA, De S, Delany ME, Dodgson JB, Dong JJ, Evans C, Frederickson KM, Flicek P, Florea L, Folkerts O, Groenen MA, Harkins TT, Herrero J, Hoffmann S, Megens HJ, Jiang A, de Jong P, Kaiser P, Kim H, Kim KW, Kim S, Langenberger D, Lee MK, Lee T, Mane S, Marcais G, Marz M, McElroy AP, Modise T, Nefedov M, Notredame C, Paton IR, Payne WS, Pertea G, Prickett D, Puiu D, Qioa D, Raineri E, Ruffier M, Salzberg SL, Schatz MC, Scheuring C, Schmidt CJ, Schroeder S, Searle SM, Smith EJ, Smith J, Sonstegard TS, Stadler PF, Tafer H, Tu ZJ, Van Tassell CP, Vilella AJ, Williams KP, Yorke JA, Zhang L, Zhang HB, Zhang X, Zhang Y, Reed KM (2010) Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol 8:e1000475
Danchin E, Vitiello V, Vienne A, Richard O, Gouret P, McDermott MF, Pontarotti P (2004) The major histocompatibility complex origin. Immunol Rev 198:216–232
Derr LK (1998) The involvement of cellular recombination and repair genes in RNA-mediated recombination in Saccharomyces cerevisiae. Genetics 148:937–945
Fink GR (1987) Pseudogenes in yeast? Cell 49:5–6
Flajnik MF, Kasahara M (2001) Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15:351–362
Flajnik MF, Kasahara M (2010) Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11:47–59
Goldberg AL (1995) Functions of the proteasome: the lysis at the end of the tunnel. Science 268:522–523
Griffin TA, Nandi D, Cruz M, Fehling HJ, Kaer LV, Monaco JJ, Colbert RA (1998) Immunoproteasome assembly: cooperative incorporation of interferon γ (IFN-γ)-inducible subunits. J Exp Med 187:97–104
Groettrup M, Kirk CJ, Basler M (2010) Proteasomes in immune cells: more than peptide producers? Nat Rev Immunol 10:73–78
Groll M, Ditzel L, Loewe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386:463–471
Hansen JD, Strassburger P, Thorgaard GH, Young WP, Du Pasquier L (1999) Expression, linkage, and polymorphism of MHC-related genes in rainbow trout, Oncorhynchus mykiss. J Immunol 163:774–786
Hayashi M, Ishibashi T, Tanaka K, Kasahara M (1997) The mouse genes encoding the third pair of β-type proteasome subunits regulated reciprocally by IFN-γ: structural comparison, chromosomal localization, and analysis of the promoter. J Immunol 159:2760–2770
Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, Dodgson JB, Chinwalla AT, Cliften PF, Clifton SW, Delehaunty KD, Fronick C, Fulton RS, Graves TA, Kremitzki C, Layman D, Magrini V, McPherson JD, Miner TL, Minx P, Nash WE, Nhan MN, Nelson JO, Oddy LG, Pohl CS, Randall-Maher J, Smith SM, Wallis JW, Yang SP, Romanov MN, Rondelli CM, Paton B, Smith J, Morrice D, Daniels L, Tempest HG, Robertson L, Masabanda JS, Griffin DK, Vignal A, Fillon V, Jacobbson L, Kerje S, Andersson L, Crooijmans RP, Aerts J, van der Poel JJ, Ellegren H, Caldwell RB, Hubbard SJ, Grafham DV, Kierzek AM, McLaren SR, Overton IM, Arakawa H, Beattie KJ, Bezzubov Y, Boardman PE, Bonfield JK, Croning MD, Davies RM, Francis MD, Humphray SJ, Scott CE, Taylor RG, Tickle C, Brown WR, Rogers J, Buerstedde JM, Wilson SA, Stubbs L, Ovcharenko I, Gordon L, Lucas S, Miller MM, Inoko H, Shiina T, Kaufman J, Salomonsen J, Skjoedt K, Wong GK, Wang J, Liu B, Yu J, Yang H, Nefedov M, Koriabine M, Dejong PJ, Goodstadt L, Webber C, Dickens NJ, Letunic I, Suyama M, Torrents D, von Mering C, Zdobnov EM, Makova K, Nekrutenko A, Elnitski L, Eswara P, King DC, Yang S, Tyekucheva S, Radakrishnan A, Harris RS, Chiaromonte F, Taylor J, He J, Rijnkels M, Griffiths-Jones S, Ureta-Vidal A, Hoffman MM, Severin J, Searle SM, Law AS, Speed D, Waddington D, Cheng Z, Tuzun E, Eichler E, Bao Z, Flicek P, Shteynberg DD, Brent MR, Bye JM, Huckle EJ, Chatterji S, Dewey C, Pachter L, Kouranov A, Mourelatos Z, Hatzigeorgiou AG, Paterson AH, Ivarie R, Brandstrom M, Axelsson E, Backstrom N, Berlin S, Webster MT, Pourquie O, Reymond A, Ucla C, Antonarakis SE, Long M, Emerson JJ, Betran E, Dupanloup I, Kaessmann H, Hinrichs AS, Bejerano G, Furey TS, Harte RA, Raney B, Siepel A, Kent WJ, Haussler D, Eyras E, Castelo R, Abril JF, Castellano S, Camara F, Parra G, Guigo R, Bourque G, Tesler G, Pevzner PA, Smit A, Fulton LA, Mardis ER, Wilson RK (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Hogquist KA, Xing Y (2010) Why CD8+ T cells need diversity when growing up. Immunity 32:5–6
Kaessmann H, Vinckenbosch N, Long M (2009) RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet 10:19–31
Kandil E, Namikawa C, Nonaka M, Greenberg AS, Flajnik MF, Ishibashi T, Kasahara M (1996) Isolation of low molecular mass polypeptide complementary DNA clones from primitive vertebrates: implications for the origin of MHC class I-restricted antigen presentation. J Immunol 156:4245–4253
Kasahara M (2007) The 2R hypothesis: an update. Curr Opin Immunol 19:547–552
Kasahara M (2010) Genome duplication and T cell immunity. Prog Mol Biol Transl Sci 92:7–36
Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T (1996) Chromosomal localization of the proteasome Z subunit gene reveals an ancient chromosomal duplication involving the major histocompatibility complex. Proc Natl Acad Sci USA 93:9096–9101
Kasahara M, Nakaya J, Satta Y, Takahata N (1997) Chromosomal duplication and the emergence of the adaptive immune system. Trends Genet 13:90–92
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kaufman J, Milne S, Gobel TW, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S (1999) The chicken B locus is a minimal essential major histocompatibility complex. Nature 401:923–925
Klein L, Hinterberger M, Wirnsberger G, Kyewski B (2009) Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol 9:833–844
Koch M, Camp S, Collen T, Avila D, Salomonsen J, Wallny HJ, van Hateren A, Hunt L, Jacob JP, Johnston F, Marston DA, Shaw I, Dunbar PR, Cerundolo V, Jones EY, Kaufman J (2007) Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity 27:885–899
Kohda K, Matsuda Y, Ishibashi T, Tanaka K, Kasahara M (1997) Structural analysis and chromosomal localization of the Psmb5 gene coding for the constitutively expressed β-type proteasome subunit. Immunogenetics 47:77–87
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Meyer A, Van de Peer Y (2005) From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27:937–945
Michalova V, Murray BW, Sultmann H, Klein J (2000) A contig map of the Mhc class I genomic region in the zebrafish reveals ancient synteny. J Immunol 164:5296–5305
Mourier T, Jeffares DC (2003) Eukaryotic intron loss. Science 300:1393
Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K (2007) Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 316:1349–1353
Murata S, Takahama Y, Tanaka K (2008) Thymoproteasome: probable role in generating positively selecting peptides. Curr Opin Immunol 20:192–196
Murray BW, Sultmann H, Klein J (1999) Analysis of a 26-kb region linked to the Mhc in zebrafish: genomic organization of the proteasome component β/tansporter associated with antigen processing-2 gene cluster and identification of five new proteasome β subunit genes. J Immunol 163:2657–2666
Nil A, Firat E, Sobek V, Eichmann K, Niedermann G (2004) Expression of housekeeping and immunoproteasome subunit genes is differentially regulated in positively and negatively selecting thymic stroma subsets. Eur J Immunol 34:2681–2689
Nitta T, Murata S, Sasaki K, Fujii H, Ripen AM, Ishimaru N, Koyasu S, Tanaka K, Takahama Y (2010) Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity 32:29–40
Ohno S (1970) Evolution by gene duplication. Springer, New York
Pamer E, Cresswell P (1998) Mechanisms of MHC class I-restricted antigen processing. Annu Rev Immunol 16:323–358
Postlethwait JH (2007) The zebrafish genome in context: ohnologs gone missing. J Exp Zool B Mol Dev Evol 308:563–577
Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutierrez EL, Dubchak I, Garcia-Fernandez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin IT, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071
Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, Prozorovski T, Lange N, Steffen J, Rieger M, Kuckelkorn U, Aktas O, Kloetzel PM, Kruger E (2010) Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 142:613–624
Shiina T, Shimizu S, Hosomichi K, Kohara S, Watanabe S, Hanzawa K, Beck S, Kulski JK, Inoko H (2004) Comparative genomic analysis of two avian (quail and chicken) MHC regions. J Immunol 172:6751–6763
Takahama Y, Nitta T, Ripen AM, Nitta S, Murata S, Tanaka K (2010) Role of thymic cortex-specific self-peptides in positive selection of T cells. Semin Immunol 22:287–293
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. doi:10.1093/molbev/msr1121
Tanaka K (2009) The proteasome: overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci 85:12–36
Tanaka K, Kasahara M (1998) The MHC class I ligand-generating system: roles of immunoproteasomes and the interferon-γ-inducible proteasome activator PA28. Immunol Rev 163:161–176
Tomaru U, Ishizu A, Murata S, Miyatake Y, Suzuki S, Takahashi S, Kazamaki T, Ohara J, Baba T, Iwasaki S, Fugo K, Otsuka N, Tanaka K, Kasahara M (2009) Exclusive expression of proteasome subunit β5t in the human thymic cortex. Blood 113:5186–5191
Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T (2002) The structure of the mammalian 20S proteasome at 2.75 Å resolution. Structure 10:609–618
Van de Peer Y, Maere S, Meyer A (2009) The evolutionary significance of ancient genome duplications. Nat Rev Genet 10:725–732
Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S (2007) Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol 5:e101
Wallny HJ, Avila D, Hunt LG, Powell TJ, Riegert P, Salomonsen J, Skjodt K, Vainio O, Vilbois F, Wiles MV, Kaufman J (2006) Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc Natl Acad Sci USA 103:1434–1439
Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, Searle S, White S, Vilella AJ, Fairley S, Heger A, Kong L, Ponting CP, Jarvis ED, Mello CV, Minx P, Lovell P, Velho TA, Ferris M, Balakrishnan CN, Sinha S, Blatti C, London SE, Li Y, Lin YC, George J, Sweedler J, Southey B, Gunaratne P, Watson M, Nam K, Backstrom N, Smeds L, Nabholz B, Itoh Y, Whitney O, Pfenning AR, Howard J, Volker M, Skinner BM, Griffin DK, Ye L, McLaren WM, Flicek P, Quesada V, Velasco G, Lopez-Otin C, Puente XS, Olender T, Lancet D, Smit AF, Hubley R, Konkel MK, Walker JA, Batzer MA, Gu W, Pollock DD, Chen L, Cheng Z, Eichler EE, Stapley J, Slate J, Ekblom R, Birkhead T, Burke T, Burt D, Scharff C, Adam I, Richard H, Sultan M, Soldatov A, Lehrach H, Edwards SV, Yang SP, Li X, Graves T, Fulton L, Nelson J, Chinwalla A, Hou S, Mardis ER, Wilson RK (2010) The genome of a songbird. Nature 464:757–762
Acknowledgments
This work was supported by grants from The Ministry of Education, Culture, Sports, Science and Technology of Japan, and National Institutes of Health grant AI027877. Yoichi Sutoh and Mizuho Kondo are supported by the Research Fellowship of the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sequence data reported in this paper have been submitted to the DDBJ/EMBL/NCBI databases under accession numbers AB624351 and AB624352.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. 1S
Amino acid sequence alignment of β5i, β5, and β5t subunits from various species. H1–H5 and β1–β12 stand for helices and β-strands, respectively. The locations of secondary structures are based on the information deposited in the RCSB Protein Data Bank. This alignment was used for the construction of the tree shown in Fig. 1. (PDF 2606 kb)
Fig. 2S
Anolis Ensembl scaffold GL344036.1 encodes MHC class I, TAP1, TAP2, and PSMB8. The MHC class I gene shown here appears to be a classical class I gene as its predicted α1 and α2 domains have eight conserved residues that are involved in the anchoring of peptide termini. The draft genome sequence contains gaps upsteam of the PSMB8 gene (indicated by a broken line) and lacks the presumed exon 1 sequence of PSMB8. (PDF 186 kb)
Rights and permissions
About this article
Cite this article
Sutoh, Y., Kondo, M., Ohta, Y. et al. Comparative genomic analysis of the proteasome β5t subunit gene: implications for the origin and evolution of thymoproteasomes. Immunogenetics 64, 49–58 (2012). https://doi.org/10.1007/s00251-011-0558-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-011-0558-0