Abstract
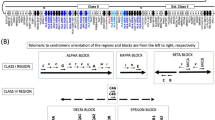
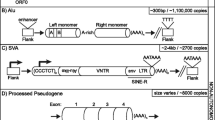
The study of the association of the Human Leukocyte Antigen (HLA) alleles and polymorphic retrotransposons such as Alu, HERV, and LTR at various loci within the Major Histocompatibility Complex allows for a better identification and stratification of disease associations and the origins of HLA haplotypes in different populations. This paper provides sequence and association data on two structurally polymorphic MER9-LTR retrotransposons that are located 54 kb apart and in close proximity to the multiallelic HLA-A gene involved in the regulation of the human immune system. Direct DNA sequencing and analysis of the PCR products identified DNA nucleotide variations between the MER9-LTR sequences at the two loci and their associations with HLA-A alleles as potential haplotype and evolutionary markers. All MER9-LTR sequences were haplotypic when associated with common HLA-A alleles. The number of SNP loci was 2.5 times greater for the solo LTR at the AK locus, which is located closer to the HLA-A gene than the solo or 3′ LTR at the HG locus. Our study shows that the nucleotide variations of the MER9-LTR DNA sequences are additional informative markers in fine mapping HLA-A genomic haplotypes for future population, evolutionary, and disease studies.


Similar content being viewed by others
References
Anzai T, Shiina T, Kimura N, Yanagiya K, Kohara S, Shigenari A, Yamagata T, Kulski JK, Naruse TK, Fujimori Y et al (2003) Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc Natl Acad Sci USA 100:7708–7713. doi:10.1073/pnas.1230533100
Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379. doi:10.1038/nrg798
Belancio VP, Hedges DJ, Deininger P (2008) Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res 18:343–358. doi:10.1101/gr.5558208
Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE (2004) Natural genetic variation caused by transposable elements in humans. Genetics 168:933–951. doi:10.1534/genetics.104.031757
Dunn DS, Inoko H, Kulski JK (2006) The association between non-melanoma skin cancer and a young dimorphic Alu element within the major histocompatibility complex class I genomic region. Tissue Antigens 68:127–134. doi:10.1111/j.1399-0039.2006.00631.x
Dunn DS, Choy MK, Phipps ME, Kulski JK (2007) The distribution of major histocompatibility complex class I polymorphic Alu insertions and their associations with HLA alleles in a Chinese population from Malaysia. Tissue Antigens 70:136–143. doi:10.1111/j.1399-0039.2007.00868.x
Gaudieri S, Dawkins RL, Habara K, Kulski JK, Gojobori T (2000) SNP profile within the human major histocompatibility complex reveals an extreme and interrupted level of nucleotide diversity. Genome Res 10:1579–1586. doi:10.1101/gr.127200
Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP (1998) Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol 9:585–598. doi:10.1006/mpev.1998.0495
Hampe A, Coriton O, Andrieux N, Carn G, Lepourcelet M, Mottier S, Dréano S, Gatius MT, Hitte C, Soriano N, Galibert F (1999) A 356-Kb sequence of the subtelomeric part of the MHC Class I region. DNA Seq 10:263–299
Horton R, Gibson R, Coggill P, Miretti M, Allcock RJ, Almeida J, Forbes S, Gilbert JG, Halls K, Harrow JL, Hart E et al (2008) Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 60:1–18. doi:10.1007/s00251-007-0262-2
Kapitonov VV, Pavlicek A, Jurka J (2004) Anthology of human repetitive DNA. In: Meyers RA (ed) Encyclopedia of Molecular Cell Biology and Molecular Medicine, vol. 1. Wiley-VCH Verlag GmbH and Co, KGaA Weinheim, pp 251–305
Kulski JK, Dunn DS (2005) Polymorphic Alu insertions within the Major Histocompatibility Complex class I genomic region. A brief review. Cytogenet Genome Res 110:193–202. doi:10.1159/000084952
Kulski JK, Gaudieri S, Inoko H, Dawkins RL (1999) Comparison between two human endogenous retrovirus (HERV)-rich regions within the major histocompatibility complex. J Mol Evol 48:675–683. doi:10.1007/PL00006511
Kulski JK, Martinez P, Longman-Jacobsen N, Wang W, Williamson J, Dawkins RL, Shiina T, Naruse T, Inoko H (2001) The association between HLA-A alleles and an Alu dimorphism near HLA-G. J Mol Evol 53:114–123. doi:10.1007/s002390010251
Kulski JK, Anzai T, Shiina T, Inoko H (2004) Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol 21:2079–2091. doi:10.1093/molbev/msh216
Kulski JK, Anzai T, Inoko H (2005) ERVK9, transposons and the evolution of MHC class I duplicons within the alpha-block of the human and chimpanzee. Cytogenet Genome Res 110:181–192. doi:10.1159/000084951
Kulski JK, Shigenari A, Shiina T, Ota M, Hosomichi K, James I, Inoko H (2008) Human endogenous retrovirus (HERVK9) structural polymorphism with haplotypic HLA-A allelic associations. Genetics 180:445–457. doi:10.1534/genetics.108.090340
Lander ES, Linton LM, Birren B et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921. doi:10.1038/35057062
Mager DL, Medstrand P (2003) Retroviral repeat sequences. In: Cooper DN (ed) Nature encyclopedia of the human genome, vol 5. Macmillan, London, pp 57–63
Mayer J, Meese E (2005) Human endogenous retroviruses in the primate lineage and their influence on host genomes. Cytogenet Genome Res 110:448–456. doi:10.1159/000084977
McKenzie L, Pecon-Slattery J, Carrington M, O'Brien S (1999) Taxonomic hierarchy of HLA class I allele sequences. Genes Immun 1:120–129. doi:10.1038/sj.gene.6363648
Medstrand P, Blomberg J (1993) Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol 67:6778–6787
Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C (2005) Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol 79:341–352. doi:10.1128/JVI.79.1.341-352.2005
Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M, Anzai T, Kulski JK, Kikkawa E, Naruse T et al (2006) Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics 173:1555–1570. doi:10.1534/genetics.106.057034
Steiper ME, Young NM (2006) Primate molecular divergence dates. Mol Phylogenet Evol 41:384–394. doi:10.1016/j.ympev.2006.05.021
Stewart CA, Horton R, Allcock RJ et al (2004) Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 14:1176–1187. doi:10.1101/gr.2188104
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Terreros MC, Martinez L, Herrera RJ (2005) Polymorphic Alu insertions and genetic diversity among African populations. Hum Biol 77:675–704. doi:10.1353/hub.2006.0009
Watanabe Y, Tokunaga K, Geraghty DE, Tadokoro K, Juji T (1997) Large-scale comparative mapping of the MHC class I region of predominant haplotypes in Japanese. Immunogenetics 46:135–141. doi:10.1007/s002510050252
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kulski, J.K., Shigenari, A., Shiina, T. et al. HLA-A allele associations with viral MER9-LTR nucleotide sequences at two distinct loci within the MHC alpha block. Immunogenetics 61, 257–270 (2009). https://doi.org/10.1007/s00251-009-0364-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-009-0364-0