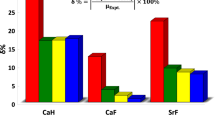
Abstract
In this review article, we discuss and analyze some recently developed hybrid atomistic–mesoscopic solvent models for multiscale biomolecular simulations. We focus on the biomolecular applications of the adaptive resolution scheme (AdResS), which allows solvent molecules to change their resolution back and forth between atomistic and coarse-grained representations according to their positions in the system. First, we discuss coupling of atomistic and coarse-grained models of salt solution using a 1-to-1 molecular mapping—i.e., one coarse-grained bead represents one water molecule—for development of a multiscale salt solution model. In order to make use of coarse-grained molecular models that are compatible with the MARTINI force field, one has to resort to a supramolecular mapping, in particular to a 4-to-1 mapping, where four water molecules are represented with one coarse-grained bead. To this end, bundled atomistic water models are employed, i.e., the relative movement of water molecules that are mapped to the same coarse-grained bead is restricted by employing harmonic springs. Supramolecular coupling has recently also been extended to polarizable coarse-grained water models with explicit charges. Since these coarse-grained models consist of several interaction sites, orientational degrees of freedom of the atomistic and coarse-grained representations are coupled via a harmonic energy penalty term. The latter aligns the dipole moments of both representations. The reviewed multiscale solvent models are ready to be used in biomolecular simulations, as illustrated in a few examples.

Adapted with permission from (Zavadlav et al. 2015b). Copyright 2015 American Chemical Society

Adapted with permission from Zavadlav et al. (2015b). Copyright 2015 American Chemical Society

Figure reprinted from Zavadlav (2015)

Figure adapted from Zavadlav et al. (2014a)

Figure adapted from Zavadlav et al. (2014a)

Reprinted from Zavadlav et al. (2015a) with the permission of AIP Publishing

Adapted from Zavadlav et al. (2015a) with the permission of AIP Publishing

Adapted from Zavadlav et al. (2015a) with the permission of AIP Publishing

Figure reprinted from Zavadlav et al. (2014b)

Figure adapted from Zavadlav et al. (2014b)

Reprinted with permission from Zavadlav et al. (2015b). Copyright 2015 American Chemical Society

Adapted with permission from Zavadlav et al. (2015b). Copyright 2015 American Chemical Society
Similar content being viewed by others
References
Agarwal A, Delle Site L (2015) Path integral molecular dynamics within the grand canonical-like adaptive resolution technique: simulation of liquid water. J Chem Phys 143:094102
Agarwal A, Delle Site L (2016) Grand-canonical adaptive resolution centroid molecular dynamics: Implementation and application. Comput Phys Commun 206:26–34
Agarwal A, Wang H, Schütte C, Delle Site L (2014) Chemical potential of liquids and mixtures via adaptive resolution simulation. J. Chem. Phys. 141:034102
Alekseevaa U, Winklerc RG, Sutmanna G (2016) Hydrodynamics in adaptive resolution particle simulations: multiparticle collision dynamics. J Comput Phys 314:14–34
Allen MP, Tildesley DJ (1989) Computer simulation of liquids. Oxford University Press, New York
Ayton GS, Noid WG, Voth GA (2007) Multiscale modeling of biomolecular systems: in serial and in parallel. Curr Opin Struct Biol 17:192–198
Bagchi B (2012) From anomalies in neat liquid to structure, dynamics and function in the biological world. Chem Phys Lett 529:1–9
Basdevant N, Borgis D, Ha-Duong T (2007) A coarse-grained protein–protein potential derived from an all-atom force field. J Phys Chem B 111:9390–9399
Bereau T, Deserno M (2009) Generic coarse-grained model for protein folding and aggregation. J Chem Phys 130:235106
Bevc S (2013) Razvoj računalniških orodij za molekularno modeliranje. PhD thesis, Faculty of Mathematics, Natural Sciences and Information Technologies, University of Primorska
Bevc S, Junghans C, Kremer K, Praprotnik M (2013) Adaptive resolution simulation of salt solutions. New J Phys 15:105007
Bevc S, Junghans C, Praprotnik M (2015) Stock: structure mapper and online coarse-graining kit for molecular simulations. J Comput Chem 36:467–477
Bock H, Gubbins KE, Klapp SH (2007) Coarse graining of nonbonded degrees of freedom. Phys Rev Lett 98:267801
Cameron A (2005) Concurrent dual-resolution Monte Carlo simulation of liquid methane. J Chem Phys 123:234101
Carmichael SP, Shell MS (2012) A new multiscale algorithm and its application to coarse-grained peptide models for self-assembly. J Phys Chem B 116:8383–8393
Crow = Columns and Rows of Workstations. http://www.cmm.ki.si/ vrana/. 28 August 2015
Chebaro Y, Pasquali S, Derreumaux P (2012) The coarse-grained OPEP force field for non-amyloid and amyloid proteins. J Phys Chem B 116:8741–8752
Chopraa G, Summab CM, Levitt M (2008) Solvent dramatically affects protein structure refinement. Proc Natl Acad Sci USA 105:20239–20244
Cragnolini T, Derreumaux P, Pasquali S (2013) Coarse-grained simulations of RNA and DNA duplexes. J Phys Chem B 117:8047–8060
Cuervo A, Dans PD, Carrascosa JL, Orozco M, Gomila G, Fumagalli L (2014) Direct measurement of the dielectric polarization properties of DNA. Proc Natl Acad Sci USA 111:3624–3630
Dans PD, Walther J, Gómez H, Orozco M (2016) Multiscale simulation of DNA. Curr Opin Chem Biol 37:29–45
Dans PD, Zeida A, Machado MR, Pantano S (2010) A coarse grained model for atomic-detailed DNA simulations with explicit electrostatics. J Chem Theory Comput 6:1711–1725
Delgado-Buscalioni R, Kremer K, Praprotnik M (2008) Concurrent triple-scale simulation of molecular liquids. J Chem Phys 128:114110
Delgado-Buscalioni R, Kremer K, Praprotnik M (2009) Coupling atomistic and continuum hydrodynamics through a mesoscopic model: application to liquid water. J Chem Phys 131:244107
Delgado-Buscalioni R, Sablić J, Praprotnik M (2015) Open boundary molecular dynamics. Eur Phys J Spec Top 224:2331–2349
Delgado-Buscalioni R, Sablić J, Praprotnik M (2015) Reply to comment by R. Klein on open boundary molecular dynamics. Eur Phys J Spec Top 224:2511–2513
Delle Site L (2016) Formulation of Liouville’s theorem for grand ensemble molecular simulations. Phys Rev E 93:022130
Delle Site L, Abrams CF, Alavi A, Kremer K (2002) Polymers near metal surfaces: selective adsorption and global conformations. Phys Rev Lett 89:156103
Duan Y, Wu C, Chowdhury S, Lee M, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012
Español P, Delgado-Buscalioni R, Everaers R, Potestio R, Donadio D, Kremer K (2015) Statistical mechanics of Hamiltonian adaptive resolution simulations. J Chem Phys 142:064115
Fabritiis GD, Delgado-Buscalioni R, Coveney PV (2006) Multiscale modeling of liquids with molecular specificity. Phys Rev Lett 97:134501
Fedosov DA, Karniadakis GE (2009) Triple-decker: interfacing atomistic–mesoscopic–continuum flow regimes. J Comput Phys 228:1157–1171
Fogarty AC, Potestio R, Kremer K (2015) Adaptive resolution simulation of a biomolecule and its hydration shell: structural and dynamical properties. J Chem Phys 142:195101
Fogarty AC, Potestio R, Kremer K (2016) A multi-resolution model to capture both global fluctuations of an enzyme and molecular recognition in the ligand-binding site. Proteins Struct Funct Bioinform 84:1902–1913
Foley T, Shell MS, Noid WG (2015) The impact of resolution upon entropy and information in coarse-grained models. J Chem Phys 143:243104
Frenkel D, Smit B (2001) Understanding molecular simulation: from algorithms to applications. Academic Press, San Diego
Fritsch S, Poblete S, Junghans C, Ciccotti G, Delle Site L, Kremer K (2012) Adaptive resolution molecular dynamics simulation through coupling to an internal particle reservoir. Phys Rev Lett 108:170602
Fuhrmans M, Sanders BP, Marrink SJ, de Vries AH (2010) Effects of bundling on the properties of the SPC water model. Theor Chem Acc 125:335–344
Gavryushov S (2008) Electrostatics of B-DNA in NaCl and CaCl\(_2\) solutions: ion size, interionic correlation, and solvent dielectric saturation effects. J Phys Chem B 112:8955–8965
Goga N, Costache S, Marrink SJ (2009) A multiscalling constant lambda molecular dynamic gromacs implementation. Mater Plast 46:53–57
Gonzales HC, Darré L, Pantano S (2013) Transferable mixing of atomistic and coarse-grain water models. J Phys Chem B 117:14438–14448
Gopal S, Mukherjee S, Cheng YM, Feig M (2010) PRIMO/PRIMONA: a coarse-grained model for proteins and nucleic acids that preserves near-atomistic accuracy. Proteins Struct Funct Bioinform 78:1266–1281
Gopal SM, Kuhn AB, Schäfer LV (2015) Systematic evaluation of bundled SPC water for biomolecular simulations. Phys Chem Chem Phys 17:8393–8406
Halverson JD, Brandes T, Lenz O, Arnold A, Bevc S, Starchenko V, Kremer K, Stuehn T, Reith D (2013) ESPResSo++: a modern multiscale simulation package for soft matter systems. Comput Phys Commun 184:1129–1149
Han W, Schulten K (2012) Further optimization of a hybrid united-atom and coarse-grained force field for folding simulations: improved backbone hydration and interactions between charged side chains. J Chem Theory Comput 8:4413–4424
Harmandarisab VA, Adhikari NP, van der Vegt NFA, Kremer K (2006) Hierarchical modeling of polystyrene: from atomistic to coarse-grained simulations. Macromolecules 39:67086719
Harmandarisab VA, Kremer K (2009) Predicting polymer dynamics at multiple length and time scales. Soft Matter 5:3920–3926
Hess B, León S, van der Vegt N, Kremer K (2006) Long time atomistic polymer trajectories from coarse grained simulations: bisphenol-A polycarbonate. Soft Matter 2:409–414
Heyden A, Truhlar DG (2008) Conservative algorithm for an adaptive change of resolution in mixed atomistic/coarse-grained multiscale simulations. J Chem Theory Comput 4:217–221
Hinckley DM, Lequieu JP, de Pablo JJ (2014) Coarse-grained modeling of DNA oligomer hybridization: length, sequence, and salt effects. J Chem Phys 141:035102
Ingólfsson HI, Lopez CA, Uusitalo JJ, de Jong DH, Gopal S, Periole X, Marrink SJ (2014) The power of coarse-graining in biomolecular simulations. WIREs Comput Mol Sci 4:225–248
Izvekov S, Parrinello M, Burnham CB, Voth GA (2004) Effective force fields for condensed phase systems from ab initio molecular dynamics simulation: a new method for force-matching. J Chem Phys 120:10896–10913
Izvekov S, Voth GA (2005) A multiscale coarse-graining method for biomolecular systems. J Phys Chem B 109:2469–2473
Izvekov S, Voth GA (2005) Multiscale coarse graining of liquid-state systems. J Chem Phys 123:134105
Izvekov S, Voth GA (2006) Multiscale coarse-graining of mixed phospholipid/cholesterol bilayers. J Chem Theory Comput 2:637648
Jedlovszky P, Vincze A, Horvai G (2007) Full description of the orientational statistics of molecules near to interfaces. Water at the interface with CCl4. Phys Chem Chem Phys 6:1874–1879
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Kamerlin SCL, Vicatos S, Dryga A, Warshel A (2011) Coarse-grained (multiscale) simulations in studies of biophysical and chemical systems. Annu Rev Phys Chem 62:41–64
Karplus M, McCammon JA (2002) Molecular dynamics simulations of biomolecules. Nat Struct Biol 9:646–652
Knotts TAI, Rathore N, Schwartz DC, de Pablo JJ (2007) A coarse grain model for DNA. J Chem Phys 126:084901
Korolev N, Lyubartsev AP, Laaksonen A, Nordenskiöld L (2006) A molecular dynamics simulation study of oriented DNA with polyamine and sodium counterions: diffusion and averaged binding of water and cations. Nucleic Acids Res 31:5971–5981
Kranenburg M, Nicolas JP, Smit B (2004) Comparison of mesoscopic phospholipid–water models. Phys Chem Chem Phys 6:4142–4151
Kreis K, Donadio D, Kremer K, Potestio R (2014) A unified framework for force-based and energy-based adaptive resolution simulations. EPL 108:30007
Kreis K, Fogarty A, Kremer K, Potestio R (2015) Advantages and challenges in coupling an ideal gas to atomistic models in adaptive resolution simulations. Eur Phys J Spec Top 224:2289–2304
Kreis K, Potestio R, Kremer K, Fogarty AC (2016) Adaptive resolution simulations with self-adjusting high-resolution regions. J Chem Theory Comput 12:4067–4081
Kuhn AB, Gopal SM, Schäfer LV (2015) On using atomistic solvent layers in hybrid all-atom/coarse-grained molecular dynamics simulations. J Chem Theory Comput 11:4460–4472
Lamm G, Pack GR (1997) Calculation of dielectric constants near polyelectrolytes in solution. J Phys Chem B 101:959–965
Lu J, Yuqing Qiu Y, Baron R, Molinero V (2014) Coarse-graining of TIP4P/2005, TIP4P-Ew, SPC/E, and TIP3P to monatomic anisotropic water models using relative entropy minimization. J Chem Theory Comput 10:4104–4120
Lyubartsev AP (2005) Multiscale modeling of lipids and lipid bilayers. Eur Biophys J 35:53–61
Lyubartsev AP, Laaksonen A (1995) Calculation of effective interaction potentials from radial distribution functions: a reverse Monte Carlo approach. Phys Rev E 52:3730–3737
Lyubartsev AP, Naômé A, Vercauteren DP, Laaksonen A (2015) Systematic hierarchical coarse-graining with the inverse Monte Carlo method. J Chem Phys 143:243120
Machado MR, Dans PD, Pantano S (2011) A hybrid all-atom/coarse grain model for multiscale simulations of DNA. Phys Chem Chem Phys 13:18134–18144
Machado MR, Pantano S (2015) Exploring Lacl–DNA dynamics by multiscale simulations using the SIRAH force field. J Chem Theory Comput 11:5012–5023
Maciejczyk M, Spasic A, Liwo A, Scheraga HA (2014) DNA duplex formation with a coarse-grained model. J Chem Theory Comput 10:5020–5035
Maffeo C, Ngo TTM, Ha T, Aksimentiev A (2014) A coarse-grained model of unstructured single-stranded DNA derived from atomistic simulation and single-molecule experiment. J Chem Theory Comput 10:2891–2896
Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The martini force field: coarse grained model for biomolecular simulations. J Phys Chem B 111:7812–7824
Masella M, Borgis D, Cuniasse P (2008) Combining a polarizable force-field and a coarse-grained polarizable solvent model: application to long dynamics simulations of bovine pancreatic trypsin inhibitor. J Comput Chem 29:1707–1724
Masella M, Borgis D, Cuniasse P (2011) Combining a polarizable force-field and a coarse-grained polarizable solvent model. II. Accounting for hydrophobic effects. J Comput Chem 32:2664–2678
Matysiak S, Clementi C, Praprotnik M, Kremer K, Delle Site L (2008) Modeling diffusive dynamics in adaptive resolution simulation of liquid water. J Chem Phys 128:024503
Michel J, Orsi M, Essex JW (2008) Prediction of partition coefficients by multiscale hybrid atomic-level/coarse-grain simulations. J Phys Chem B 112:657–660
Mohamed KM, Mohamad AA (2010) A review of the development of hybrid atomistic-continuum methods for dense fluids. Microfluid Nanofluid 8:283–302
Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ (2008) The MARTINI coarse grained force field: extension to proteins. J Chem Theory Comput 4:819–834
Mukherji D, Kremer K (2013) Coil–globule–coil transition of pnipam in aqueous methanol: coupling all-atom simulations to semi-grand canonical coarse-grained reservoir. Macromolecules 46:9158–9163
Mullinax JW, Noid WG (2009) Extended ensemble approach for deriving transferable coarse-grained potentials. J Chem Phys 131:104110
Nagarajan A, Junghans C, Matysiak S (2013) Multiscale simulation of liquid water using a four-to-one mapping for coarse-graining. J Chem Theory Comput 9:5168–5175
Neri M, Anselmi C, Cascella M, Maritan A, Carloni P (2005) Coarse-grained model of proteins incorporating atomistic detail of the active site. Phys Rev Lett 95:218102
Neumann M (1983) Dipole-moment fluctuation formulas in computer-simulations of polar systems. Mol Phys 50:841–858
Neumann M (1985) The dielectric constant of water. Computer simulations with the MCY potential. J Chem Phys 82:5663–5672
Nielsen SO, Moore PB, Ensing B (2010) Adaptive multiscale molecular dynamics of macromolecular fluids. Phys Rev Lett 105:237802
Noid WG (2013) Perspective: Coarse-grained models for biomolecular systems. J Chem Phys 139:090901
Orsi M, Ding W, Palaiokostas M (2014) Direct mixing of atomistic solutes and coarse-grained water. J Chem Theory Comput 10:4684–4693
Orsi M, Essex JW (2011) The ELBA force field for coarse-grain modeling of lipid membranes. PLoS One 6:e28637
Ouldridge TE, Louis AA, Doye JPK (2011) Structural, mechanical and thermodynamic properties of a coarse-grained DNA model. J Chem Phys 134:085101
Periole X, Marrink SJ (2013) The MARTINI coarse-grained force field. In: Monticelli L, Salonen E (eds) Biomolecular simulations: methods and protocols, methods in molecular biology, vol 924. Springer, New York, pp 533–565
Peter C, Kremer K (2009) Multiscale simulation of soft matter systems—from the atomistic to the coarse-grained level and back. Soft Matter 5:4357–4366
Peters JH, Klein R, Delle Site L (2016) Simulation of macromolecular liquids with the adaptive resolution molecular dynamics technique. Phys Rev E 94:023309
Poblete S, Praprotnik M, Kremer K, Delle Site L (2010) Coupling different levels of resolution in molecular simulations. J Chem Phys 132:114101
Poma AB, Delle Site L (2010) Classical to path-integral adaptive resolution in molecular simulation: towards a smooth quantum-classical coupling. Phys Rev Lett 104:250201
Poma AB, Delle Site L (2011) Adaptive resolution simulation of liquid para-hydrogen: testing the robustness of the quantum-classical adaptive coupling. Phys Chem Chem Phys 13:10510–10519
Potestio R, Español P, Delgado-Buscalioni R, Everaers R, Kremer K, Donadio D (2013) Monte Carlo adaptive resolution simulation of multicomponent molecular liquids. Phys Rev Lett 111:060601
Potestio R, Fritsch S, Español P, Delgado-Buscalioni R, Kremer K, Everaers R, Donadio D (2013) Hamiltonian adaptive resolution simulation for molecular liquids. Phys Rev Lett 110:108301
Potestio R, Peter C, Kremer K (2014) Computer simulations of soft matter: linking the scales. Entropy 16:4199–4245
Potoyan DA, Savelyev A, Papoian GA (2013) Recent successes in coarse-grained modeling of DNA. WIREs Comput Mol Sci 3:69–83
Praprotnik M, Delle Site L, Kremer K (2005) Adaptive resolution molecular-dynamics simulation: changing the degrees of freedom on the fly. J Chem Phys 123:224106
Praprotnik M, Delle Site L, Kremer K (2008) Multiscale simulation of soft matter: from scale bridging to adaptive resolution. Annu Rev Phys Chem 59:545–571
Praprotnik M, Matysiak S, Delle Site L, Kremer K, Clementi C (2007) Adaptive resolution simulation of liquid water. J Phys Condens Matter 19:292201
Praprotnik M, Poblete S, Kremer K (2011) Statistical physics problems in adaptive resolution computer simulations of complex fluids. J Stat Phys 145:946–966
Reith D, Pütz M, Müller-Plathe F (2003) Deriving effective mesoscale potentials from atomistic simulations. J Comput Chem 24:1624–1636
Reynwar BJ, Illya G, Harmandaris VA, Müller MM, Kremer K, Deserno M (2007) Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447:461
Riniker S, Eichenberger AP, van Gunsteren WF (2012) Solvating atomic level fine-grained proteins in supra-molecular level coarse-grained water for molecular dynamics simulations. Eur Biophys J 41:647–661
Riniker S, Eichenberger AP, van Gunsteren WF (2012) Structural effects of an atomic-level layer of water molecules around proteins solvated in supra-molecular coarse-grained water. J Phys Chem B 116:8873–8879
Riniker S, van Gunsteren WF (2011) A simple, efficient polarizable coarse-grained water model for molecular dynamics simulations. J Chem Phys 134:084110
Rudzinski JF, Noid WG (2015) Bottom–up coarse-graining of peptide ensembles and helixcoil transitions. J Chem Theory Comput 11:1278–1291
Rzepiela AJ, Louhivuori M, Peter C, Marrink SJ (2011) Hybrid simulations: combining atomistic and coarse-grained force fields using virtual sites. Phys Chem Chem Phys 13:10437–10448
Sablić J, Praprotnik M, Delgado-Buscalioni R (2016) Open boundary molecular dynamics of sheared star-polymer melts. Soft Matter 12:2416–2439
Savelyev A, Papoian GA (2010) Chemically accurate coarse graining of double-stranded DNA. Proc Natl Acad Sci USA 107:20340–20345
Shaw DE, Deneroff MM, Dror RO, Kuskin JS, Larson RH, Salmon JK, Young C, Batson B, Bowers KJ, Chao JC, Eastwood MP, Gagliardo J, Grossman JP, Ho CR, Ierardi DJ, Kolossváry I, Klepeis JL, Layman T, McLeavey C, Moraes MA, Mueller R, Priest EC, Shan Y, Spengler J, Theobald M, Towles B, Wang SC (2007) Anton, a special-purpose machine for molecular dynamics simulation. SIGARCH Comput Archit News 35:1–12
Shell MS (2008) The relative entropy is fundamental to thermodynamic ensemble optimization. J Chem Phys 129:144108
Shelley JC, Shelley MY, Reeder R, Bandyopadhyay S, Klein ML (2001) A coarse grained model for phospholipid simulations. J Phys Chem B 105:4464–4470
Shen L, Hu H (2014) Resolution-adapted all-atomic and coarse-grained model for biomolecular simulations. J Chem Theory Comput 10:2528–2536
Shen L, Yang W (2016) Quantum mechanics/molecular mechanics method combined with hybrid all-atom and coarse-grained model: Theory and application on Redox potential calculations. J Chem Theory Comput. doi:10.1021/acs.jctc.5b01107
Shi Q, Izvekov S, Voth GA (2006) Mixed atomistic and coarse-grained molecular dynamics: simulation of a membrane-bound ion channel. J Phys Chem B 110:15045–15048
Snodin BEK, Randisi F, Mosayebi M, Sulc P, Schreck JS, Romano F, Ouldridge TE, Tsukanov R, Nir E, Louis AA, Doye JPK (2015) Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J Chem Phys 142:234901
Sokkar P, Boulanger E, Thiel W, Sanchez-Garcia E (2015) Hybrid quantum mechanics/molecular mechanics/coarse grained modeling: a triple-resolution approach for biomolecular systems. J Chem Theory Comput 11:1809–1818
Sokkar P, Choi SM, Rhee YM (2013) Simple method for simulating the mixture of atomistic and coarse-grained molecular systems. J Chem Theory Comput 9:3728–3739
Stanley C, Rau D (2011) Evidence for water structuring forces between surfaces. Curr Opin Colloid Interface Sci 16:551–556
Tironi IG, Sperb R, Smith PE, van Gunsteren WF (1995) A generalized reaction field method for molecular dynamics simulations. J Chem Phys 102:5451–5459
Tschöp W, Kremer K, Hahn O, Batoulis J, Bürger T (1998) Simulation of polymer melts. II. From coarse-grained models back to atomistic description. Acta Polym 49:75–79
Tuckerman ME (2010) Statistical mechanics: theory and molecular simulation. Oxford University Press, New York
Uusitalo JJ, Ingólfsson HI, Akhshi P, Tieleman DP, Marrink SJ (2015) Martini coarse-grained force field: extension to DNA. J Chem Theory Comput 11:3932–3945
Villa E, Balaeff A, Mahadevan L, Schulten K (2004) Multiscale method for simulating protein–DNA complexes. Multiscale Model Simul 2:527–553
Villa E, Balaeff A, Schulten K (2005) Structural dynamics of the lac repressor–DNA complex revelead by a multiscale simulation. Proc Natl Acad Sci USA 102:6783–6788
Walther JH, Praprotnik M, Kotsalis EM, Koumoutsakos P (2012) Multiscale simulation of water flow pas a C540 fullerene. J Comput Phys 231:2677–2681
Wang H, Agarwal A (2015) Adaptive resolution simulation in equilibrium and beyond. Eur Phys J Spec Top 224:2269–2287
Wang H, Hartmann C, Schütte C, Delle Site L (2013) Grand-canonical-like molecular-dynamics simulations by using an adaptive-resolution technique. Phys Rev X 3:011018
Wang ZJ, Deserno M (2010) A systematically coarse-grained solvent-free model for quantitative phospholipid bilayer simulations. J Phys Chem B 114:11207–11220
Wassenaar TA, Ingólfsson HI, Böckmann RA, Peter Tieleman DP, Marrink SJ (2015) Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput 11:2144–2155
Wassenaar TA, Ingólfsson HI, Priess M, Marrink SJ, Schaefer LV (2013) Mixing martini: electrostatic coupling in hybrid atomistic–coarse-grained biomolecular simulations. J Phys Chem B 117:3516–3530
Yesylevskyy SO, Schäfer LV, Sengupta D, Marrink SJ (2010) Polarizable water model for the coarse-grained MARTINI force field. PLoS Comput Biol 6:e1000810
Young MA, Jayaram B, Beveridge DL (1998) Local dielectric environment of B-DNA in solution: results from a 14 ns molecular dynamics trajectory. J Phys Chem B 102:7666–7669
Zavadlav J (2015) Multiscale simulation of biomolecular systems. PhD thesis, Department of Physics, Faculty of Mathematics and Physics, University of Ljubljana
Zavadlav J, Marrink SJ, Praprotnik M (2016) Adaptive resolution simulation of supramolecular water: the concurrent making, breaking, and remaking of water bundles. J Chem Theory Comput 12:4138–4145
Zavadlav J, Melo MN, Cunha AV, de Vries AH, Marrink SJ, Praprotnik M (2014) Adaptive resolution simulation of martini solvents. J Chem Theory Comput 10:2591–2598
Zavadlav J, Melo MN, Marrink SJ, Praprotnik M (2014) Adaptive resolution simulation of an atomistic protein in martini water. J Chem Phys 140:054114
Zavadlav J, Melo MN, Marrink SJ, Praprotnik M (2015) Adaptive resolution simulation of polarizable supramolecular coarse-grained water models. J Chem Phys 142:244118
Zavadlav J, Podgornik R, Melo MN, Marrink SJ, Praprotnik M (2016) Adaptive resolution simulation of an atomistic DNA molecule in MARTINI salt solution. Eur Phys J Spec Top 225:1595–1607
Zavadlav J, Podgornik R, Praprotnik M (2015) Adaptive resolution simulation of a DNA molecule in salt solution. J Chem Theory Comput 11:5035–5044
Zhou HX (2014) Theoretical frameworks for multiscale modeling and simulation. Curr Opin Struct Biol 25:67–76
Acknowledgements
We would like to thank S. J. Marrink and M. N. Melo for a fruitful collaboration on coupling atomistic and MARTINI molecular models for biomolecular simulations. We are grateful to C. Junghans and K. Kremer for collaboration on the salt solution. We would also like to thank R. Podgornik for collaborating with us on the DNA simulations and J. Sablić for careful reading of the manuscript. We acknowledge financial support through grants P1-0002 and J1-7135 from the Slovenian Research Agency.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: Regional Biophysics Conference 2016.
Rights and permissions
About this article
Cite this article
Zavadlav, J., Bevc, S. & Praprotnik, M. Adaptive resolution simulations of biomolecular systems. Eur Biophys J 46, 821–835 (2017). https://doi.org/10.1007/s00249-017-1248-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-017-1248-0