Abstract
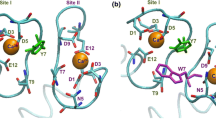
The paper reports the results of numerical calculations of the pKa’s of the ionizable groups and the electrostatic interactions between calmodulin lobes in three different states of calmodulin: calcium-free, peptide-free; calcium-loaded, peptide-free; and calcium-loaded, peptide-bound. NMR and X-ray studies revealed that in these states the overall structure of calmodulin adopts various conformations referred as: disordered semi-compact, extended and compact conformations, respectively. In addition, a new X-ray structure was recently reported (Structure, 2003, 11, 1303) showing that calcium-loaded, peptide-free calmodulin can also adopt a compact conformation in addition to the well known extended conformation. The calculated energy changes of calcium-loaded, peptide-free calmodulin along the pathway connecting these two conformations provide a possible explanation for this structural plasticity. The effect of pH and organic compounds in the solution phase on the preference of calmodulin to adopt compact or extended conformations may be thus rationalized. Analysis of the contribution of the ionization changes to the energy of association of calmodulin lobes suggested that the formation of the compact forms requires protonation of several acidic residues. However, two different protonation scenarios are revealed: a protonation due to internal lobe organization and thus independent of the lobes association, and a protonation induced by the lobes association resulting to a proton uptake. In addition, the role of the individual residues on the energy of association of calmodulin lobes is calculated in two compact conformations (peptide-free and peptide-bound) and is shown that a set of residues always plays a dominant role in inter-domain interactions.






Similar content being viewed by others
Abbreviations
- CaM:
-
Calmodulin
- CFPF:
-
Calcium-free, peptide-free
- CLPF:
-
Calcium-loaded, peptide-free
- CLPB:
-
Calcium-loaded, peptide-bound
References
Alexandrov N, Shindyalov I (2003) PDP: protein domain parser. Bioinformatics 19:429–430
Alexandrov V, Lehnert U, Echols N, Milburn D, Engelman D, Gerstein M (2005) Normal modes for predicting protein motions: a comprehensive database assessment and associated Web tool. Protein Sci 14:633–643
Alexov E (2003) The role of the protein side chain fluctuations on the strength of pair wise electrostatic interactions. Comparing experimental with computed pKa’s. Proteins 50:94–103
Alexov E (2004a) Calculating proteon uptake/release and the binding free energy taking into account ionization and conformation changes induced by protein-inhibitor association. Application to plasmepsin, cathepsin D and endothiapepsin-pepstatin complexes. Proteins 56:572–584
Alexov E (2004b) Numerical calculations of the pH of maximal protein stability. The effect of the sequence composition and 3D structure. Eur J Biochem 271:173–185
Alexov E, Gunner M (1997) Incorporating protein conformation flexibility into the calculation of pH-dependent protein properties. Biophys J 74:2075–2093
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Bhattacharya S, Bunick C, Chazin W (2004) Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta (BBA)—Mol Cell Res 1742:69–79
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4:187–217
Broxk R, Scheek R, Weljie A, Vogel H (2004) Backbone dynamic properties of the central linker region of calcium-calmodulin in 35% trifluroethanol. J Struct Biol 146:272–280
Chattopadhyaya R, Meador WE, Means AR, Quiocho FA (1992) Calmodulin structure refined at 1.7 A resolution. J Mol Biol 228:1177–1192
Fallon J, Quiocho F (2003) A closed compact structure of native Ca2+-Calmodulin. Structure 11:1303–1307
Felts AK, Harano Y, Gallicchio E, Levy RM (2004) Free energy surfaces of beta-hairpin and alpha-helical peptides generated by replica exchange molecular dynamics with the AGBNP implicit solvent model. Proteins 56:310–321
Forrest L, Honig B (2005) An assessment of the accuracy of methods for predicting hydrogen positions in protein structures. Proteins 61:296–309
Georgescu R, Alexov E, Gunner M (2002) Combining conformational flexibility and continuum electrostatics for calculating residue pKa’s in proteins. Biophys J 83:1731–1748
Grabarek Z (2005) Structure of a trapped intermediate of calmodulin: calcium regulation of EF-hand proteins from a new perspective. J Mol Biol 346:1351–1366
Hoeflich K, Ikura M (2002) Calmodulinin action: diversity in target recognition and activation mechanisms. Cell 108:739–742
Hultschiga C, Hecht H, Frank R (2004) Systematic delineation of a calmodulin peptide interaction. J Mol Biol 343:559–568
Ikura M (1996) Calcium binding and conformational response in EF-hand proteins. TIBS 21:14–17
Kesvatera T, Jonsson B, Thulin E, Linse S (2001) Focusing of the electrostatic potential at EF-hands of calbindin D(9k): titration of acidic residues. Proteins 45:129–135
Krebs W, Gerstein M (2000) The morph server: a standartized system for analyzing and visualizing macromolecular motionsin a database framework. Nucleic Acids Res 28:1665–1675
Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A (1995) Solution structure of calcium-free calmodulin. Nat Struct Biol 2:768–779
MacKerell AD Jr, Bashford D, Bellot M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem 102:3586–3616
Meador WE, Means AR, Quiocho FA (1992) Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin–peptide complex. Science 257:1251–1254
Nicholls A, Sharp KA, Honig H (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281–296
Ohashi I, Pohoreki R, Morita K, Stemmer P (2004) Alcohols increase calmodulin affinity for Ca2+ and decrease target affinity for calmodulin. Biochem Biophys Acta 1691:161–167
Persechini A, Kretsinger R (1988) The central helix of calmodulin functions as a flexible tether. J Biol Chem B 101:12175–12178
Petrey D, Honig B (2003) “GRASP2: visualization, surface properties and electrostatic of macromolecular structures. Methods Enzymol 374:492–509
Ponder JW (1999) TINKER-software tools for molecular design. Washington University, St. Louis
Rocchia W, Alexov E, Honig B (2001) Extending the applicability of the nonlinear Poisson–Boltzmann equation: multiple dielectric constants and multivalent ions. J Phys Chem 105:6507–6514
Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B (2002) Rapid grid-based construction of the molecular surface and the use of induced surface charges to calculate reaction field energies: applications to the molecular systems and geometrical objects. J Comput Chem 23:128–137
Seedher N, Bhatia S (2003) Solubility enhancement of Cox-2 inhibitors using various solvent systems. AAPS PharmSciTech 4:1–9
Sitkoff D, Sharp KA, Honig B (1994) Accurate calculation of hydration free energies using macroscopic solvent models. J Phys Chem 98:1978–1988
Slaughter B, Bieber-Urbauer R, Johnson C (2005a) Single-molecule tracking of sub-millisecond domain motion in calmodulin. J Phys Chem B 109(26):12658–12662
Slaughter B, Unruh J, Allen M, Bieber R, Johnson C (2005b) Conformational substates of calmodulin revealed by single-pair fluorescence resonance energy transfer: influence of solution conditions and oxidative modification. Biochemistry 44:3694–3707
Strynadka N, James M (1989) Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem 58:951–998
Uchikoga N, Takahashi SY, Ke R, Sonoyama M, Mitaku S (2005) Electric charge balance mechanism of extended soluble proteins. Protein Sci 14:74–80
Yang C, Jas GS, Kuczera K (2004) Structure, dynamics and interaction with kinase targets: computer simulations of calmodulin. Biochim Biophys Acta 1697:289–300
Zhang M, Tanaka T, Ikura M (1995) Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat Struct Biol 2:758–767
Zhu J, Alexov E, Honig B (2005) Comparative study of generalized born models: born radii and peptide folding. J Phys Chem B 109:3008–3022
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Isvoran, A., Craescu, C.T. & Alexov, E. Electrostatic control of the overall shape of calmodulin: numerical calculations. Eur Biophys J 36, 225–237 (2007). https://doi.org/10.1007/s00249-006-0123-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00249-006-0123-1