Abstract
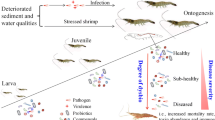
It is now recognized that some diseases of aquatic animals are attributed to polymicrobial pathogens infection. Thus, the traditional view of “one pathogen, one disease” might mislead the identification of multiple pathogens, which in turn impedes the design of probiotics. To address this gap, we explored polymicrobial pathogens based on the origin and timing of increased abundance over shrimp white feces syndrome (WFS) progression. OTU70848 Vibrio fluvialis, OTU35090 V. coralliilyticus, and OTU28721 V. tubiashii were identified as the primary colonizers, whose abundances increased only in individuals that eventually showed disease signs but were stable in healthy subjects over the same timeframe. Notably, the random Forest model revealed that the profiles of the three primary colonizers contributed an overall 91.4% of diagnosing accuracy of shrimp health status. Additionally, NetShift analysis quantified that the three primary colonizers were important “drivers” in the gut microbiotas from healthy to WFS shrimp. For these reasons, the primary colonizers were potential pathogens that contributed to the exacerbation of WFS. By this logic, we further identified a few “drivers” commensals in healthy individuals, such as OUT50531 Demequina sediminicola and OTU_74495 Ruegeria lacuscaerulensis, which directly antagonized the three primary colonizers. The predicted functional pathways involved in energy metabolism, genetic information processing, terpenoids and polyketides metabolism, lipid and amino acid metabolism significantly decreased in diseased shrimp compared with those in healthy cohorts, in concordant with the knowledge that the attenuations of these functional pathways increase shrimp sensitivity to pathogen infection. Collectively, we provide an ecological framework for inferring polymicrobial pathogens and designing antagonized probiotics by quantifying their changed “driver” feature that intimately links shrimp WFS progression. This approach might generalize to the exploring disease etiology for other aquatic animals.






Similar content being viewed by others
References
Espín JC (2017) Gut microbiota, diet and health. Mol Nutr Food Res 61:1770015
Li T, Tian D, Zhu Z et al (2019) The gut microbiota: a new perspective on the toxicity of malachite green (MG). Appl Microbiol Biotechnol 103:23–24
Xiong J, Dai W, Zhu J, Liu K, Dong C, Qiu Q (2017) The underlying ecological processes of gut microbiota among cohabitating retarded, overgrown and normal shrimp. Microb Ecol 73:988–999
Hou D, Huang Z, Zeng S, Liu J, Weng S, He J (2018) Comparative analysis of the bacterial community compositions of the shrimp intestine, surrounding water and sediment. J Appl Microbiol 125:792–799
Zhu J, Dai W, Qiu Q, Dong C, Zhang J, Xiong J (2016) Contrasting ecological processes and functional compositions between intestinal bacterial community in healthy and diseased shrimp. Microb Ecol 72:975–985
Cornejo-Granados F, Gallardo-Becerra L, Leonardo-Reza M et al (2018) A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ 6:e5382
Yang G, Tian X, Dong S, Liang L (2019) Bacillus cereus and rhubarb regulate the intestinal microbiota of sea cucumber (Apostichopus japonicus Selenka): species-species interaction, network, and stability. Aquaculture 512:734284
Weng FH, Shaw GW, Weng CY et al (2017) Inferring microbial interactions in the gut of the Hong Kong whipping frog (Polypedates megacephalus) and a validation using probiotics. Front Microbiol 8:525
Vonaesch P, Anderson M, Sansonetti PJ (2018) Pathogens, microbiome and the host: emergence of the ecological Koch’s postulates. FEMS Microbiol Rev 42:273–292
Rogers GB, Hoffman LR, Carroll MP, Bruce KD (2013) Interpreting infective microbiota: the importance of an ecological perspective. Trends Microbiol 21:271–276
Gignoux-Wolfsohn SA, Aronson FM, Vollmer SV (2017) Complex interactions between potentially pathogenic, opportunistic, and resident bacteria emerge during infection on a reef-building coral. FEMS Microbiol Ecol 93:fix080
Stephens WZ, Burns AR, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ (2016) The composition of the zebrafish intestinal microbial community varies across development. ISME J 10:644–654
Burns AR, Stephens WZ, Stagaman K, Wong S, Rawls JF, Guillemin K, Bohannan BJ (2016) Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J 10:655–664
Xiong J, Dai W, Qiu Q, Zhu J, Yang W, Li C (2018) Response of host–bacterial colonization in shrimp to developmental stage, environment and disease. Mol Ecol 27:3686–3699
Dai W, Chen J, Xiong J (2019) Concept of microbial gatekeepers: positive guys? Appl Microbiol Biotechnol 103:633–641
Montoya JM, Pimm SL, Solé RV (2006) Ecological networks and their fragility. Nature 442:259
Banerjee S, Schlaeppi K, vander Heijden MGA (2018) Keystone taxa as drivers of microbiome structure and functioning. Nat Rev Microbiol 16:567–576
Nicholson JK, Holmes E, Kinross J et al (2012) Host-gut microbiota metabolic interactions. Science (80- ) 336:1262–1267
Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE, Flint HJ (2006) Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599
Round JL, Mazmanian SK (2009) The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323
Buffie CG, Bucci V, Stein RR, McKenney P, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink M, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG (2015) Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 517:205–208
Dai W, Yu W, Xuan L, et al (2018) Integrating molecular and ecological approaches to identify potential polymicrobial pathogens over a shrimp disease progression. Appl Microbiol Biotechnol 102:3755–3764
Coyte KZ, Schluter J, Foster KR (2015) The ecology of the microbiome: networks, competition, and stability. Science 350:663–666
Deng Y, Jiang YH, Yang Y et al (2012) Molecular ecological network analyses. BMC Bioinformatics 13:113
Kuntal BK, Chandrakar P, Sadhu S, Mande SS (2019) ‘NetShift’: a methodology for understanding ‘driver microbes’ from healthy and disease microbiome datasets. ISME J 13:442–454
Sriurairatana S, Boonyawiwat V, Gangnonngiw W et al (2014) White feces syndrome of shrimp arises from transformation, sloughing and aggregation of hepatopancreatic microvilli into vermiform bodies superficially resembling gregarines. PLoS One 9:e99170
Xiong J, Zhu J, Dai W, Dong C, Qiu Q, Li C (2017) Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ Microbiol 19:1490–1501
Selvin J, Huxley AJ, Lipton AP (2004) Immunomodulatory potential of marine secondary metabolites against bacterial diseases of shrimp. Aquaculture. 230:241–248
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Caporaso JG, Bittinger K, Bushman FD et al (2009) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267
Oksanen J, Blanchet FG, Kindt R, et al (2010) Vegan: community ecology package. R package version 1.17–4. http//cran r-project/org
Harrell FE, Harrell Jr MFE (2019) Package ‘Hmisc.’ CRAN2018 235–236
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345–366
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2:18–22
Xiong J (2018) Progress in the gut microbiota in exploring shrimp disease pathogenesis and incidence. Appl Microbiol Biotechnol 102:7343–7350
Ben-Haim Y, Thompson FL, Thompson CC, Cnockaert MC, Hoste B, Swings J, Rosenberg E (2003) Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 53:309–315
Ramayo-Caldas Y, Mach N, Lepage P, Levenez F, Denis C, Lemonnier G, Leplat JJ, Billon Y, Berri M, Doré J, Rogel-Gaillard C, Estellé J (2016) Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME J 10:2973–2977
Xiong J, Wang K, Wu J et al (2015) Changes in intestinal bacterial communities are closely associated with shrimp disease severity. Appl Microbiol Biotechnol 99:6911–6919
Clemente JC, Ursell LK, Laura Wegener P, Rob K (2012) The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270
Huang Z, Li X, Wang L, Shao Z (2016) Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquac Res 47:1737–1746
Yao Z, Yang K, Huang L et al (2018) Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Express 8:120
Wang J, Huang Y, Xu K, Zhang X, Sun H, Fan L, Yan M (2019) White spot syndrome virus (WSSV) infection impacts intestinal microbiota composition and function in Litopenaeus vannamei. Fish Shellfish Immunol 84:130–137
Yu W, Wu JH, Zhang J et al (2018) A meta-analysis reveals universal gut bacterial signatures for diagnosing the incidence of shrimp disease. FEMS Microbiol Ecol 94:fiy147
Phumkhachorn P, Rattanachaikunsopon P (2010) Isolation and partial characterization of a bacteriophage infecting the shrimp pathogen Vibrio harveyi. Afr J Microbiol Res 4:1794–1800
Xiong J, Dai W, Li C (2016) Advances, challenges, and directions in shrimp disease control: the guidelines from an ecological perspective. Appl Microbiol Biotechnol 100:6947–6954
Igbinosa E, Okoh AI (2010) Vibrio fluvialis: an unusual enteric pathogen of increasing public health concern. Int J Environ Res Public Health 7:3628–3643
Garren M, Son K, Raina JB, Rusconi R, Menolascina F, Shapiro OH, Tout J, Bourne DG, Seymour JR, Stocker R (2014) A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J 8:999–1007
Hamilton AL, Kamm MA, Ng SC, Morrison M (2018) Proteus spp. as putative gastrointestinal pathogens. Clin Microbiol Rev 31:e00085–e00017
Sandaruwan Kumara KRP, Hettiarachchi M (2017) White faeces syndrome caused by vibrio alginolyticus and vibrio fluvialis in shrimp, Penaeus monodon (Fabricius 1798)-multimodal strategy to control the syndrome in Sri Lankan grow-out ponds. Asian Fish Sci 30:245–261
Silvester R, Alexander D, George M, Hatha AAM (2017) Prevalence and multiple antibiotic resistance of Vibrio coralliilyticus, along the southwest coast of India. Curr Sci 112:1749–1755
Lightner DV (2011) Virus diseases of farmed shrimp in the Western hemisphere (the Americas): a review. J Invertebr Pathol 106:110–130
Mallon CA, Van Elsas JD, Salles JF (2015) Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol 23:719–729
Jones SE, Lennon JT (2010) Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci U S A 107:5881–5886
Bressan W (2003) Biological control of maize seed pathogenic fungi by use of actinomycetes. BioControl 48:233–240
Newaj-Fyzul A, Al-Harbi AH, Austin B (2014) Developments in the use of probiotics for disease control in aquaculture. Aquaculture 431:1–11
Xiong J, Zhu J, Zhang D (2014) The application of bacterial indicator phylotypes to predict shrimp health status. Appl Microbiol Biotechnol 98:8291–8299
Douglas GM, Beiko RG, Langille MGI (2018) Predicting the functional potential of the microbiome from marker genes using PICRUSt. In: Beiko R, Hsiao W, Parkinson J (eds) Microbiome Analysis. Methods in Molecular Biology, Humana Press, New York, 1849:169–17
Immanuel G, Sivagnanavelmurugan M, Palavesam A (2011) Antibacterial effect of medium-chain fatty acid: caprylic acid on gnotobiotic Artemia franciscana nauplii against shrimp pathogens Vibrio harveyi and V. parahaemolyticus. Aquac Int 19:91–101
Funding
This work was supported by the National Natural Science Foundation of China (31872693), the Natural Science Fund for Distinguished Young Scholars of Zhejiang Province (LR19C030001), the Technology Innovation Team of Ningbo (2015C110018), and the K.C. Wong Magna Fund in Ningbo University.
Author information
Authors and Affiliations
Contributions
JC and JX conceived and designed research. JL, XZ, and QQ conducted experiments. JX contributed to analytical tools. JL and JX analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
This article does not contain any studies with human participants performed by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of shrimp followed the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Additional information
Key points
• Polymicrobial pathogens were identified by distinguishing shrimp age from disease effect.
• Potential pathogens were validated by “driver” feature and diagnosis accuracy.
• Taxa that directly antagonized the candidate pathogens were identified.
• We exemplified the identification of polymicrobial pathogens and the design of antagonized probiotics.
Electronic Supplementary Material
ESM 1
(DOC 3492 kb)
Rights and permissions
About this article
Cite this article
Lu, J., Zhang, X., Qiu, Q. et al. Identifying Potential Polymicrobial Pathogens: Moving Beyond Differential Abundance to Driver Taxa. Microb Ecol 80, 447–458 (2020). https://doi.org/10.1007/s00248-020-01511-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-020-01511-y