Abstract
In the present study, we assessed prokaryotic communities of demosponges, a calcareous sponge, octocorals, sediment and seawater in coral reef habitat of the central Red Sea, including endemic species and species new to science. Goals of the study were to compare the prokaryotic communities of demosponges with the calcareous sponge and octocorals and to assign preliminary high microbial abundance (HMA) or low microbial abundance (LMA) status to the sponge species based on compositional trait data. Based on the compositional data, we were able to assign preliminary LMA or HMA status to all sponge species. Certain species, however, had traits of both LMA and HMA species. For example, the sponge Ectyoplasia coccinea, which appeared to be a LMA species, had traits, including a relatively high abundance of Chloroflexi members, that were more typical of HMA species. This included dominant OTUs assigned to two different classes within the Chloroflexi. The calcareous sponge clustered together with seawater, the known LMA sponge Stylissa carteri and other presumable LMA species. The two dominant OTUs of this species were assigned to the Deltaproteobacteria and had no close relatives in the GenBank database. The octocoral species in the present study had prokaryotic communities that were distinct from sediment, seawater and all sponge species. These were characterised by OTUs assigned to the orders Rhodospirillales, Cellvibrionales, Spirochaetales and the genus Endozoicomonas, which were rare or absent in samples from other biotopes.




Similar content being viewed by others
References
Plaisance L, Caley MJ, Brainard RE, Knowlton N (2011) The diversity of coral reefs: what are we missing? PLoS One 6:e25026. https://doi.org/10.1371/journal.pone.0025026
Hughes TP, Graham NAJ, Jackson JBC, Mumby PJ, Steneck RS (2010) Rising to the challenge of sustaining coral reef resilience. Trends Ecol Evol 25:633–642
Done TJ, DeVantier LM, Turak E, Fisk DA, Wakeford M, van Woesik R (2010) Coral growth on three reefs: development of recovery benchmarks using a space for time approach. Coral Reefs 29:815–833
Cleary DFR, Polónia ARM, Renema W, Hoeksema BW, Wolstenholme J, Tuti Y, de Voogd NJ (2014) Coral reefs next to a major conurbation: a study of temporal change (1985-2011) in coral cover and composition in the reefs of Jakarta Indonesia. Mar Ecol Prog Ser 501:89–98. https://doi.org/10.3354/meps10678
Cleary DFR, Polónia ARM, Renema W, Hoeksema BW, Rachello-Dolmen PG, Moolenbeek RG, Budiyanto A, Yahmantoro TY, Giyanto DSG, Prud'homme van Reine WF, Hariyanto R, Gittenberger A, Rikoh MS, de Voogd NJ (2016) Variation in the composition of corals, fishes, sponges, echinoderms, ascidians, molluscs, foraminifera and macroalgae across a pronounced in-to-offshore environmental gradient in the Jakarta Bay-Thousand Islands coral reef complex. Mar Pollut Bull 110:701–717. https://doi.org/10.1016/j.marpolbul.2016.04.042
Cleary DFR (2017) Linking fish species traits to environmental conditions in the Jakarta Bay Pulau Seribu coral reef system. Mar Pollut Bull 122:259–262. https://doi.org/10.1016/j.marpolbul.2017.06.054
Hentschel U, Usher KM, Taylor MW (2006) Marine sponges as microbial fermenters. FEMS Microbiol Ecol 55:167–177
Taylor MW, Radax R, Steger D, Wagner M (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295–347
Preston CM, Wu KY, Molinski TF, DeLong EF (1996) A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. Nov., sp. nov. Proc Natl Acad Sci U S A 93:6241–6246
Rohwer F, Breitbart M, Jara J, Azam F, Knowlton N (2001) Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20:85–91
Hentschel U, Hopke J, Horn M et al (2002) Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol 68:4431–4440
Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F (2007) Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9:2707–2719
Schmitt S, Deines P, Behnam F, Wagner M, Taylor MW (2011) Chloroflexi bacteria are more diverse, abundant, and similar in high than in low microbial abundance sponges. FEMS Microbiol Ecol 78:497–510. https://doi.org/10.1111/j.1574-6941.2011.01179.x
Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N, Perez T, Rodrigo A, Schupp PJ, Vacelet J, Webster N, Hentschel U, Taylor MW (2012) Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J 6:564–576
Cleary DFR, Becking LE, Pires ACC, de Voogd NJ, Egas C, Gomes NCM (2013) Habitat and host related variation in sponge bacterial communities in Indonesian coral reefs and marine lakes. FEMS Microbiol Ecol 85:465–482
de Voogd NJ, Cleary DFR, Polónia ARM, Gomes NCM (2015) Bacterial communities of four different biotopes and their functional genomic nitrogen signature from the thousand-island reef complex, West-Java, Indonesia. FEMS Microbiol Ecol 91:fiv019. https://doi.org/10.1093/femsec/fiv019
Polónia ARM, Cleary DFR, Freitas R, de Voogd NJ, Gomes NCM (2015) The putative functional ecology and distribution of archaeal communities in an Indonesian coral reef environment. Mol Ecol 24:409–423. https://doi.org/10.1111/mec.13024
van de Water JAJM, Allemand D, Ferrier-Pagès C (2018) Host-microbe interactions in octocoral holobionts—recent advances and perspectives. Microbiome 6:64. https://doi.org/10.1186/s40168-018-0431-6
Bell J (2008) The functional roles of marine sponges. Estuar Coast Shelf Sci 79:341–353. https://doi.org/10.1016/j.ecss.2008.05.002
Wulff J (2001) Assessing and monitoring coral reef sponges: why and how? Bull Mar Sci 69:831–846
Maldonado M, Aguilar R, Bannister RJ, Bell D, Conway KW, Dayton PK, Díaz C, Gutt J, Kelly M, Kenchington ELR, Leys SP, Pomponi SA, Rapp HT, Rutzler K, Tendal OS, Vacelet J, Young CM (2016) Sponge grounds as key marine habitats: a synthetic review of types, structure, functional roles, and conservation concerns. In: Rossi S, Bramanti L, Gori A, Orejas C (eds) Marine animal forests: the ecology of benthic biodiversity hotspots. Springer, Berlin
Piel J (2009) Metabolites from symbiotic bacteria. Nat Prod Rep 26:338–362
Vacelet J, Donadey C (1977) Electron-microscope study of association between spme sponges and bacteria. J Exp Mar Biol Ecol 30:301–314
Weisz JB, Hentschel U, Lindquist N, Martens CS (2007) Linking abundance and diversity of sponge-associated microbial communities to metabolic differences in host sponges. Mar Biol 152:475–483
Hochmuth T, Niederkrüger H, Gernert C, Siegl A, Taudien S, Platzer M, Crews P, Hentschel U, Piel J (2010) Linking chemical and microbial diversity in marine sponges: possible role for poribacteria as producers of methyl-branched fatty acids. Chembiochem 11:2572–2578. https://doi.org/10.1002/cbic.201000510
Thacker RW, Freeman CJ (2012) Sponge-microbe symbioses: recent advances and new directions. Adv Mar Biol 62:57–111
Moitinho-Silva L, Bayer K, Cannistraci CV, Giles EC, Ryu T, Seridi L, Ravasi T, Hentschel U (2014) Specificity and transcriptional activity of microbiota associated with low and high microbial abundance sponges from the Red Sea. Mol Ecol 23:1348–1363
Bayer K, Kamke J, Hentschel U (2014) Quantification of bacterial and archaeal symbionts in high and low microbial abundance sponges using real-time PCR. FEMS Microbol Ecol 89:679–690
Moitinho-Silva L, Steinert G, Nielsen S, Hardoim CCP, Wu YC, McCormack G, López-Legentil S, Marchant R, Webster N, Thomas T, Hentschel U (2017) Predicting the HMA-LMA status in marine sponges by machine learning. Front Microbiol 8:752. https://doi.org/10.3389/fmicb.2017.00752
Bayer K, Moitinho-Silva L, Brummer F, Cannistraci CV, Ravasi T, Hentschel U (2014) GeoChip-based insights into the microbial functional gene repertoire of marine sponges (high microbial abundance, low microbial abundance) and seawater. FEMS Microbiol Ecol 90:832–843. https://doi.org/10.1111/1574-6941.12441
Montalvo NF, Mohamed NM, Enticknap JJ, Hill RT (2005) Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 87:29–36
Montalvo NF, Davis J, Vicente J, Pittiglio R, Ravel J, Hill RT (2014) Integration of culture-based and molecular analysis of a complex sponge-associated bacterial community. PLoS One 9:e90517
Montalvo NF, Hill RT (2011) Sponge-associated bacteria are strictly maintained in two closely related but geographically distant sponge hosts. Appl Environ Microbiol 77:7207–7216
Swierts T, Cleary DFR, de Voogd NJ (2018) Biogeography of prokaryote communities in closely related giant barrel sponges across the Indo-Pacific. FEMS Microbiol Ecol 94:fiy194. https://doi.org/10.1093/femsec/fiy194
Roué M, Domart-Coulon I, Ereskovsky A, Djediat C, Perez T, Bourguet-Kondracki ML (2010) Cellular localization of clathridimine, an antimicrobial 2-aminoimidazole alkaloid produced by the Mediterranean calcareous sponge Clathrina clathrus. J Nat Prod 73:1277–1282
Quévrain E, Roué M, Domart-Coulon I, Bourguet-Kondracki ML (2014) Assessing the potential bacterial origin of the chemical diversity in calcareous sponges. J Mar Sci Technol 22:36–49
Gibbons JB, Salvant JM, Vaden RM, Kwon KH, Welm BE, Looper RE (2015) Synthesis of Naamidine A and Selective access to N2-Acyl-2-aminoimidazole analogues. J Organomet Chem 80:10076–10085
Koswatta PB, Kasiri S, Das JK, Bhan A, Lima HM, Garcia-Barboza B, Khatibi NN, Yousufuddin M, Mandal SS, Lovely CJ (2017) Total synthesis and cytotoxicity of Leucetta alkaloids. Bioorg Med Chem 25:1608–1621
Daly M, Brugler MR, Cartwright P, Collins AG, Dawson MN, Fautin DG, France SC, McFadden CS, Opresko DM, Rodriguez E, Romano SL, Stake JL (2007) The phylum Cnidaria: a review of phylogenetic patterns and diversity 300 years after Linnaeus. Zootaxa 1668:127–182
Appeltans W, Ahyong ST, Anderson G, Angel MV, Artois T, Bailly N, Bamber R, Barber A, Bartsch I, Berta A, Błażewicz-Paszkowycz M, Bock P, Boxshall G, Boyko CB, Brandão SN, Bray RA, Bruce NL, Cairns SD, Chan TY, Cheng L, Collins AG, Cribb T, Curini-Galletti M, Dahdouh-Guebas F, Davie PJ, Dawson MN, de Clerck O, Decock W, de Grave S, de Voogd NJ, Domning DP, Emig CC, Erséus C, Eschmeyer W, Fauchald K, Fautin DG, Feist SW, Fransen CH, Furuya H, Garcia-Alvarez O, Gerken S, Gibson D, Gittenberger A, Gofas S, Gómez-Daglio L, Gordon DP, Guiry MD, Hernandez F, Hoeksema BW, Hopcroft RR, Jaume D, Kirk P, Koedam N, Koenemann S, Kolb JB, Kristensen RM, Kroh A, Lambert G, Lazarus DB, Lemaitre R, Longshaw M, Lowry J, Macpherson E, Madin LP, Mah C, Mapstone G, McLaughlin P, Mees J, Meland K, Messing CG, Mills CE, Molodtsova TN, Mooi R, Neuhaus B, Ng PK, Nielsen C, Norenburg J, Opresko DM, Osawa M, Paulay G, Perrin W, Pilger JF, Poore GC, Pugh P, Read GB, Reimer JD, Rius M, Rocha RM, Saiz-Salinas JI, Scarabino V, Schierwater B, Schmidt-Rhaesa A, Schnabel KE, Schotte M, Schuchert P, Schwabe E, Segers H, Self-Sullivan C, Shenkar N, Siegel V, Sterrer W, Stöhr S, Swalla B, Tasker ML, Thuesen EV, Timm T, Todaro MA, Turon X, Tyler S, Uetz P, van der Land J, Vanhoorne B, van Ofwegen L, van Soest R, Vanaverbeke J, Walker-Smith G, Walter TC, Warren A, Williams GC, Wilson SP, Costello MJ (2012) The magnitude of global marine species diversity. Curr Biol 22:2189–2202
Fabricius K, Alderslade P (2001) Soft corals and sea fans. A comprehensive guide to the tropical shallow-water genera of the central-West Pacific, the Indian Ocean and the Red Sea. Australian Institute of Marine Science, Australia
Fabricius K, De’ath G (2008) Photosynthetic symbionts and energy supply determine octocoral biodiversity in coral reefs. Ecology 89:3163–3173
Gabay Y, Benayahu Y, Fine M (2013) Does elevated pCO2 affect reef octocorals? Ecol Evol 3:465–473
Sorokin YI (1991) Biomass, metabolic rates and feeding of some common reef zoantharians and octocorals. Aust J Mar Freshwat Res 42:729–741
Newman SP, Meesters EH, Dryden CS, Williams SM, Sanchez C, Mumby PJ, Polunin NVC (2015) Reef flattening effects on total richness and species responses in the Caribbean. J Anim Ecol 84:1678–1689
La Rivière M, Garrabou J, Bally M (2015) Evidence for host specificity among dominant bacterial symbionts in temperate gorgonian corals. Coral Reefs 34:1087–1098
van de Water JAJM, Melkonian R, Voolstra CR, Junca H, Beraud E, Allemand D, Ferrier-Pagès C (2017) Comparative assessment of Mediterranean gorgonian-associated microbial communities reveals conserved core and locally variant bacteria. Microb Ecol 73:466–478
Berumen ML, Hoey AS, Bass WH, Bouwmeester J, Catania D, Cochran JEM, Khalil MT, Miyake S, Mughal MR, Spaet JLY, Saenz-Agudelo P (2013) The status of coral reef ecology research in the Red Sea. Coral Reefs 32:737–748
Ngugi DK, Antunes A, Brune A, Stingl U (2012) Biogeography of pelagic bacterioplankton across an antagonistic tempera- ture-salinity gradient in the Red Sea. Mol Ecol 21:388–405
ReFuGe 2020 Consortium (2015) the ReFuGe 2020 Consortium-using “omics” approaches to explore the adaptability and resilience of coral holobionts to environmental change. Front Mar Sci 2:68
Gloeckner V, Wehrl M, Moitinho-Silva L, Gernert C, Schupp P, Pawlik JR, Lindquist NL, Erpenbeck D, Wörheide G, Hentschel U (2014) The HMA-LMA dichotomy revisited: an electron microscopical survey of 56 sponge species. Biol Bull 227:78–88
Cleary DFR, de Voogd NJ, Polónia ARM, Freitas R, Gomes NCM (2015) Composition and predictive functional analysis of bacterial communities in seawater, sediment and sponges in an Indonesian coral reef environment. Microb Ecol 70:889–903. https://doi.org/10.1007/s00248-015-0632-5
van Soest RWM, de Voogd NJ (2018) Calcareous sponges of the Western Indian Ocean and Red Sea. Zootaxa 4426(1):1. https://doi.org/10.11646/zootaxa.4426.1.1
Khalil MT, Bouwmeester J, Berumen ML (2017) Spatial variation in coral reef fish and benthic communities in the central Saudi Arabian Red Sea. PeerJ 5:e3410. https://doi.org/10.7717/peerj.3410
Giles EC, Saenz-Agudelo P, Hussey NE, Ravasi T, Berumen ML (2015) Exploring seascape genetics and kinship in the reef sponge Stylissa carteri in the Red Sea. Ecol Evol 5:2487–2502. https://doi.org/10.1002/ece3.1511
Kandler NM, Wooster MK, Leray M, Knowlton N, de Voogd NJ, Paulay G, Berumen ML (2018) Hyperdiverse macrofauna communities associated with a common sponge, Stylissa carteri, shift across ecological gradients in the central Red Sea. Diversity 11:18. https://doi.org/10.3390/d11020018
Roberts MB, Jones GP, McCormick MI, Munday PL, Neale S, Thorrold S, Robitzch VS, Berumen ML (2016) Homogeneity of coral reef communities across 8 degrees of latitude in the Saudi Arabian Red Sea. Mar Pollut Bull 105:558–565
Racault MF, Raitsos DE, Berumen ML, Brewin RJW, Platt T, Sathyendranath S, Hoteit I (2015) Phytoplankton phenology indices in coral reef ecosystems: application to ocean-colour observations in the Red Sea. Remote Sens Environ 160:222–234. https://doi.org/10.1016/j.rse.2015.01.019
Furby KA, Bouwmeester J, Berumen ML (2013) Susceptibility of central Red Sea corals during a major bleaching event. Coral Reefs 32:505–513
Monroe AA, Ziegler M, Roik A et al (2018) In situ observations of coral bleaching in the central Saudi Arabian Red Sea during the 2015/2016 global coral bleaching event. PLoS One 13:e0195814
Cleary DFR, Becking LE, Polónia ARM, Freitas R, Gomes NCM (2016) Jellyfish associated microbiomes of Indonesian marine lakes. FEMS Microbiol Ecol 92:fiw064. https://doi.org/10.1093/femsec/fiw064
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. https://doi.org/10.1093/nar/gks808
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Coelho FJRC, Cleary DFR, Gomes NC, Pólonia ARM, Huang YM, Liu LL, de Voogd NJ (2018) Sponge prokaryote communities in Taiwanese coral reef and shallow hydrothermal vent ecosystems. Microb Ecol 75:239–254
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
R Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria 3–900051–07-0 (Available from hyperink). http://www.R-project.org. http://www.R-project.org
Oksanen J, Guillaume Blanchet F, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2019) Vegan: Community Ecology Package. R package version 2.5-6. https://CRAN.R-project.org/package=vegan
Legendre P, Gallagher ED (2001) Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280. https://doi.org/10.1007/s004420100716
Cleary DFR (2003) An examination of scale of assessment, logging and ENSO-induced fires on butterfly diversity in Borneo. Oecologia 135:313–321. https://doi.org/10.1007/s00442-003-1188-5
de Voogd NJ, Cleary DFR, Hoeksema BW, Noor A, van Soest RWM (2006) Sponge beta diversity in the Spermonde archipelago, Indonesia. Mar Ecol Prog Ser 309:131–142
Tweedie, MCK (1984) An index which distinguishes between some important exponential families. In: Statistics: applications and New Directions-Proceedings of the Indian Statistical Institute Golden Jubilee International Conference, (eds) Ghosh JK and Roy J, Pp. 579–604. Calcutta: Indian Statistical Institute
Lenth R (2017) emmeans: estimated marginal means, aka least-squares means. https://CRAN.R project.org/package=emmeans
Swierts T, Peijnenburg KT, de Leeuw CA, Breeuwer JA, Cleary DFR, de Voogd NJ (2017) Globally intertwined evolutionary history of giant barrel sponges. Coral Reefs 36:933–945
Erwin PM, Coma R, López-Sendino P, Serrano E, Ribes M (2015) Stable symbionts across the HMA–LMA dichotomy: low seasonal and interannual variation in sponge-associated bacteria from taxonomically diverse hosts. FEMS Microbiol Ecol 91:fiv115
Cleary DFR, Swierts T, Coelho FJRC, Polónia ARM, Huang YM, Ferreira MRS, Putchakarn S, Carvalheiro L, van der Ent E, Gomes NCM, de Voogd NJ (2019) The sponge microbiome within the greater coral reef microbial metacommunity. Nat Commun 10:1644
de Voogd NJ, Polónia ARM, Gauvin-Bialecki A, Cleary DFR (2018) Assessing the microbial communities of high microbial abundance (HMA) and low microbial abundance (LMA) sponges inhabiting the remote western Indian Ocean island of Mayotte. Mar Ecol 39:e12517. https://doi.org/10.1111/maec.12517
Bayer K, Jahn MT, Slaby BM, Moitinho-Silva L, Hentschel U (2018) Marine sponges as Chloroflexi hot spots: genomic insights and high-resolution visualization of an abundant and diverse symbiotic clade. mSystems 3:e00150–e00118. https://doi.org/10.1128/mSystems.00150-18
Bryant DA, Frigaard NU (2006) Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol 14:488–496
Mehrshad M, Rodriguez-Valera F, Amoozegar MA, López-García P, Ghai R (2018) The enigmatic SAR202 cluster up close: shedding light on a globally distributed dark ocean lineage involved in sulfur cycling. ISME J 12:655–668
Cleary DFR, Polónia ARM, Becking LE, de Voogd NJ, Purwanto GH, Gomes NCM (2018) Compositional analysis of bacterial communities in seawater, sediment and high and low microbial abundance sponges in the Misool coral reef system, Indonesia. Mar Biodivers 48:1889–1901. https://doi.org/10.1007/s12526-017-0697-0
Giles EC, Kamke J, Moitinho-Silva L, Taylor MW, Hentschel U, Ravasi T, Schmitt S (2013) Bacterial community profiles in low microbial abundance sponges. FEMS Microbiol Ecol 83:232–241. https://doi.org/10.1111/j.1574-6941.2012.01467.x
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Off S, Alawi M, Spieck E (2010) Enrichment and physiological characterization of a novel Nitrospira-like bacterium obtained from a marine sponge. Appl Environ Microbiol 76:4640–4646
Offre P, Spang A, Schleper C (2013) Archaea in biogeochemical cycles. Annu Rev Microbiol 67. https://doi.org/10.1146/annurev-micro-092412-155614
Kamke J, Taylor MW, Schmitt S (2010) Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J 4:498–508. https://doi.org/10.1038/ismej.2009.143
Poppell E, Weisz J, Spicer L, Massaro A, Hill A, Hill M (2014) Sponge heterotrophic capacity and bacterial community structure in high and low microbial abundance sponges. Mar Ecol 35:414–424
Ribes M, Dziallas C, Coma R, Riemann L (2015) Microbial diversity and putative diazotrophy in high- and low-microbial-abundance mediterranean sponges. Appl Environ Microbiol 81:5683–5693. https://doi.org/10.1128/AEM.01320-15
Easson CG, Thacker RW (2014) Phylogenetic signal in the community structure of host-specific microbiomes of tropical marine sponges. Front Microbiol 5:532
Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U (2008) Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol 74:7694–7708. https://doi.org/10.1128/AEM.00878-08
Leite DCA, Leão P, Garrido AG, Lins U, Santos HF, Pires DO, Castro CB, van Elsas J, Zilberberg C, Rosado AS, Peixoto RS (2017) Broadcast spawning coral Mussismilia hispida can vertically transfer its associated bacterial core. Front Microbiol 8:176. https://doi.org/10.3389/fmicb.2017.00176
Webster NS, Thomas T (2016) The sponge Hologenome. MBio 7:e00135–e00116. https://doi.org/10.1128/mBio.00135-16
Adair KL, Douglas AE (2017) Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol 35:23–29. https://doi.org/10.1016/j.mib.2016.11.002
De Roy K, Marzorati M, Negroni A, Thas O, Balloi A, Fava F, Verstraete W, Daffonchio D, Boon N (2013) Environmental conditions and community evenness determine the outcome of biological invasion. Nat Commun 4:1383. https://doi.org/10.1038/ncomms2392
Mallon CA, Poly F, Le Roux X et al (2015) Resource pulses can alleviate the biodiversity–invasion relationship in soil microbial communities. Ecology 96:915
Eisenhauer N, Schulz W, Scheu S, Jousset A (2013) Niche dimensionality links biodiversity and invasibility of microbial communities. Funct Ecol 27:282–288
Vivant A, Garmyn D, Maron PA, Nowak V, Piveteau P (2013) Microbial diversity and structure are drivers of the biological barrier effect against Listeria monocytogenes in soil. PLoS One 8:e76911
Sheik CS, Jain S, Dick GJ (2014) Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol 16:304–317. https://doi.org/10.1111/1462-2920.12165
Cao H, Chunming D, Bougouffa S, Li J, Zhang W, Shao Z, Bajic VB, Qian PY (2016) Delta-proteobacterial SAR324 group in hydrothermal plumes on the South id-Atlantic Ridge. Sci Rep 6:22842. https://doi.org/10.1038/srep22842
Haroon MF, Thompson LR, Stingl U (2016) Draft genome sequence of uncultured SAR324 bacterium lautmerah10, binned from a Red Sea metagenome. Genome Announc 4:e01711–e01715
Bayer T, Arif C, Ferrier-Pagès C, Zoccola D, Aranda M, Voolstra C (2013) Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar Ecol Prog Ser 479:75–84
McCauley EP, Haltli B, Correa H, Kerr RG (2016) Spatial and temporal investigation of the microbiome of the Caribbean octocoral Erythropodium caribaeorum. FEMS Microbiol Ecol 92:fiw147
Neave MJ, Michell CT, Apprill A et al (2014) Whole-genome sequences of three symbiotic Endozoicomonas bacteria. Genome Announc 2:e00802–e00814
Neave MJ, Apprill A, Ferrier-Pagès C, Voolstra CR (2016) Diversity and function of prevalent symbiotic marine bacteria in the genus Endozoicomonas. Appl Microbiol Biotechnol 100:8315–8324
van de Water JAJM, Melkonian R, Junca H, Voolstra CR, Reynaud S, Allemand D, Ferrier-Pagès C (2016) Spirochaetes dominate the microbial community associated with the red coral Corallium rubrum on a broad geographic scale. Sci Rep 6:27277
La Rivière M, Roumagnac M, Garrabou J, Bally M (2013) Transient shifts in bacterial communities associated with the temperate gorgonian Paramuricea clavata in the northwestern Mediterranean Sea. PLoS One 8:e57385
Robertson V, Haltli B, McCauley E, Overy D, Kerr R (2016) Highly variable bacterial communities associated with the Octocoral Antillogorgia elisabethae. Microorganisms 4:23
Oh H-M, Kang I, Ferriera S, Giovannoni SJ, Cho J-C (2010) Genome sequence of the oligotrophic marine Gammaproteobacterium HTCC2143, isolated from the Oregon coast. J Bacteriol 192:4530–4531
Spring S, Riedel T (2013) Mixotrophic growth of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine gammaproteobacteria is carbon-starvation independent and correlates with the type of carbon source and oxygen availability. BMC Microbiol 13:117
van de Water JAJM, Voolstra CR, Rottier C, Cocito S, Peirano A, Allemand D, Ferrier-Pagès C (2018) Seasonal stability in the microbiomes of temperate gorgonians and the red coral Corallium rubrum across the Mediterranean Sea. Microb Ecol 75:1–15
Holm JB, Heidelberg KB (2016) Microbiomes of Muricea californica and M. fruticosa: comparative analyses of two co-occurring eastern Pacific octocorals. Front Microbiol 7:917
Lawler SN, Kellogg CA, France SC, Clostio RW, Brooke SD, Ross SW (2016) Coral-associated bacterial diversity is conserved across two deep-sea Anthothela species. Front Microbiol 7:458
Wessels W, Sprungala S, Watson S-A, Miller DJ, Bourne DG (2017) The microbiome of the octocoral Lobophytum pauciflorum: minor differences between sexes and resilience to short-term stress. FEMS Microbiol Ecol 93:fix013
Leadbetter JR, Schmidt TM, Graber JR, Breznak JA (1999) Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686–689
Lilburn TG, Kim KS, Ostrom NE, Byzek KR, Leadbetter JR, Breznak JA (2001) Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495–2498
Acknowledgements
We would like to thank Michael Campbell for providing the map. We are grateful for support in the field from the King Abdullah University of Science and Technology, including support from the Coastal and Marine Resources Core Lab.
Funding
This work is a contribution to the LESS CORAL project [PTDC/AAC-AMB/115304/2009] funded by FCT – Fundação para a Ciência e a Tecnologia , I.P., through national funds, and co-funding by FEDER, within the PT2020 Partnership Agreement and Compete 2020. Thanks are also due, for financial support to CESAM (UID/AMB/50017/2019), to FCT/MEC through national funds, and co-funding by the FEDER. Michael L. Berumen was supported by funding from KAUST (award no. CRG-1-814 2012-BER-002) and baseline research funds. Ana R.M. Polónia was supported by a postdoctoral scholarship (SFRH/BPD/117563/2016) funded by FCT, Portugal and national funds MCTES.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethical Approval
This article does not contain any studies with animals performed by any of the authors.
Electronic Supplementary Material
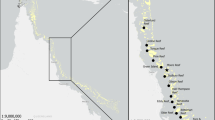
Supplementary Fig. 1
Location of the sampling area in the Red Sea. (PDF 857 kb)
Supplementary Fig. 2
Underwater photographs of all species sampled in the present study: (A) Xestospongia testudinaria, (B) Chalinula sp., (C) Petrosia elephantotus, (D) Topsentia aqabaensis, (E) Negombata magnifica, (F) Ectyoplasia coccinea, (G) Stylissa carteri, (H) Cinachyrella sp., (I) Dactylospongia metachromia, (J) Hyrtios sp., (K) Aplysinella rhax, (L) Leucetta chagosensis, (M) Ptilocaulis sp. (N) Luffariella variabilis, (O) Siphonogorgia sp. and (P) Melithaea rubrinodis. (PNG 10320 kb)
Supplementary Fig. 3
Ordination showing the third and fourth axes of the principal coordinates analysis (PCO) of OTU composition. In fig. 4a, light grey symbols represent operational taxonomic unit (OTU) scores with the symbol size proportional to their abundance (number of sequence reads). In fig. 4b, the colour of the symbol indicates the taxonomic affiliation. HMA sponges: Cs - Chalinula sp., Dm - Dactylospongia metachromia, Hy - Hyrtios sp., Lv - Luffariella variabilis, Pe - Petrosia elephantotus and Xt - Xestospongia testudinaria, LMA sponges: Ar - Aplysinella rhax, Ec - Ectyoplasia coccinea, Nm - Negombata magnifica, Cn - Cinachyrella sp., Ps - Ptilocaulis sp.., Sc - Stylissa carteri and Ta - Topsentia aqabaensis, calcareous sponge: Lc - Leucetta chagosensis, octocorals: Mr - Melithaea rubrinodis and Sg - Siphonogorgia sp., Sd - sediment and Wt – seawater. The third and fourth axes explain an additional 14% of the variation in composition. (PDF 1266 kb)
Supplementary Fig. 4
Relative abundance of the most abundant phyla in the demosponges Cs - Chalinula sp., Dm - Dactylospongia metachromia, Hy - Hyrtios sp., Lv - Luffariella variabilis, Pe - Petrosia elaphantotus and Xt - Xestospongia testudinaria, Ar - Aplysinella rhax, Ec - Ectyoplasia coccinea, Nm - Negombata magnifica, Pt - Cinachyrella sp., Ps - Ptilocaulis sp., Sc - Stylissa carteri and Ta - Topsentia aqabaensis, the calcareous sponge: Lc - Leucetta chagosensis, octocorals: Mr - Melithaea rubrinodis and Sg - Siphonogorgia sp., Sd - sediment and Wt - seawater (PDF 10 kb)
Supplementary Fig. 5
Rarefied richness of the demosponge species Cs - Chalinula sp., Dm - Dactylospongia metachromia, Hy - Hyrtios sp., Hr - Luffariella variabilis, Pe - Petrosia elaphantotus and Xt - Xestospongia testudinaria, Ar - Aplysinella rhax, Ec - Ectyoplasia coccinea, Nm - Negombata magnifica, Pt - Cinachyrella sp., Ps - Ptilocaulis sp., Sc - Stylissa carteri and Ta - Topsentia aqabaensis, the calcareous sponge: Lc - Leucetta chagosensis, the octocorals: Mr - Melithaea rubrinodis and Sg - Siphonogorgia sp., Sd - sediment and Wt - seawater. (PDF 111 kb)
Supplementary Table 1
List of samples used in the present study including the biotope, species and location. The relative percentages of the most abundant phyla, classes and orders are given as is the dominance (Dom; relative abundance of the most abundant OTU), Dom3 (relative abundance of the three most abundant OTUs), rarefied richness (Richness), Pielou’s J (J) and Shannon’s H′ diversity index (H). NCBI SRA metadata includes library_ID, study, Bioproject, accession, Biosample, seq_meth, library_strategy, library_source, library_layout, investigation_type, title, organism, project_name, lat_lon, geo_loc_name, collection_date, biome, feature, material and env_package. (XLS 98 kb)
Supplementary Table 2
Number of sequences and OTUs recorded for all phyla in the present study based on the Silva 128 database. (XLS 11 kb)
Supplementary Table 3
The number of phyla, orders and classes recorded in each biotope. (XLS 7 kb)
Supplementary Table 4
Results of emmeans analysis showing pairwise comparisons of differences in the relative abundances of selected phyla between biotopes based on the Tukey test. Significance: * 0.01 < Pr < 0.05 ** 0.001 < Pr < 0.01; *** Pr < 0.001. (XLS 468 kb)
Supplementary Table 5
Results of simper analysis showing the contribution of OTUs to differences in similarity between pairs of samples. OTUs that contribute significantly to differences are indicated: * 0.01 < P < 0.05 ** 0.001 < P < 0.01; *** P < 0.001. (XLS 1381 kb)
Supplementary Table 6
List of abundant (≥ 2500 sequence reads) OTUs and closely related organisms identified using BLAST search. OTU: OTU number; Sum: number of sequence reads; Acc: Genbank accession numbers of closely related organisms identified using BLAST; Seq: sequence similarity of these organisms with our representative OTU sequences; Source: isolation source of organisms identified using BLAST. (XLS 18 kb)
Rights and permissions
About this article
Cite this article
Cleary, D.F.R., Polónia, A., Reijnen, B. et al. Prokaryote Communities Inhabiting Endemic and Newly Discovered Sponges and Octocorals from the Red Sea. Microb Ecol 80, 103–119 (2020). https://doi.org/10.1007/s00248-019-01465-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01465-w