Abstract
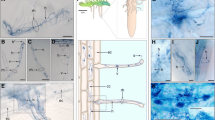
Ectomycorrhizal (ECM) fine roots account for a substantial proportion of forest production and their decomposition releases large amounts of nutrients to the soil ecosystem. However, little is known about the fungi involved in ECM decomposition, including assemblages of fungal saprotrophs, endophytes, and the ECM fungi themselves. To follow fungal succession during the degradation of senescing fine roots, understory seedlings of Abies balsamea and Picea rubens at two sites in the Acadian forest of Nova Scotia were either severed at the root collar or left as controls. Root systems were collected sequentially over two growing seasons and assessed for fine root loss and ECM mantle integrity. ECM were identified by ITS-PCR and grouped into broad morphological categories. Fungal communities colonizing the senescing fine roots were also monitored by systematically constructing clone libraries over the course of the experiment. ECM with cottony, weakly pigmented mantles (e.g., Cortinarius) degraded within the first year. Those with cottony, but intensely pigmented mantles (Piloderma), and smooth mantles with weak pigmentation (Russulaceae) degraded more slowly. Smooth, melanized ECM (Cenococcum and Tomentella) generally maintained integrity over the course of the experiment. Rates of fine root loss and changes in ECM mantle integrity were positively correlated with soil temperature. ECM DNA was detected throughout the experiment, and was not replaced by that of saprotrophic species during the two seasons sampled. However, fungal root endophytes (e.g., Helotiaceae) initially increased in abundance and then decreased as mantles degraded, suggesting a possible role in ECM decomposition.




Similar content being viewed by others
References
Jackson R, Mooney HA, Schulze ED (1997) A global budget for fine root biomass, surface area, and nutrient contents. Proc Natl Acad Sci U S A 94:7362–7366
McCormack ML, Dickie IA, Eissenstat DM, Fahey TJ, Fernandez CW, Guo D, Helmisaari HS, Hobbie EA, Iversen CM, Jackson RB, Leppälammi-Kujansuu J (2015) Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New Phytol 207:505–518
Godbold DL, Hoosbeek MR, Lukac M, Cotrufo MF, Janssens IA, Ceulemans R, Polle A, Velthorst EJ, Scarascia-Mugnozza G, De Angelis P, Miglietta F (2006) Mycorrhizal hyphal turnover as a dominant process for carbon input into soil organic matter. Plant Soil 281:15–24
Ekblad A, Wallander H, Godbold DL, Cruz C, Johnson D, Baldrian P, Björk RG, Epron D, Kieliszewska-Rokicka B, Kjøller R, Kraigher H, Matzner E, Neumann J, Plassard C (2013) The production and turnover of extramatrical mycelium of ectomycorrhizal fungi in forest soils: role in carbon cycling. Plant Soil 366:1–27
King JS, Albaugh TJ, Allen HL, Buford M, Strain BR, Dougherty P (2002) Below-ground carbon input to soil is controlled by nutrient availability and fine root dynamics in loblolly pine. New Phytol 154:389–398
Langley AJ, Chapman SK, Hungate BA (2006) Ectomycorrhizal colonization slows root decomposition: the post-mortem fungal legacy. Ecol Lett 9:955–959
Koide RT, Fernandez CW, Peoples MS (2011) Can ectomycorrhizal colonization of Pinus resinosa roots affect their decomposition? New Phytol 191:508–514
Koide RT, Malcolm GM (2009) N concentration controls decomposition rates of different strains of ectomycorrhizal fungi. Fungal Ecol 2:197–202
Fernandez CW, McCormack ML, Hill JM, Pritchard SG, Koide RT (2013) On the persistence of Cenococcum geophilum ectomycorrhizas and its implications for forest carbon and nutrient cycles. Soil Biol Biochem 65:141–143
Fernandez CW, Koide RT (2014) Initial melanin and nitrogen concentrations control the decomposition of ectomycorrhizal fungal litter. Soil Biol Biochem 77:150–157
Sterner O, Bergman R, Kihlberg J, Wickberg B (1985) The sesquiterpenes of Lactarius vellereus and their role in a proposed chemical defense system. J Nat Prod 48:279–288
Böllmann J, Elmer M, Wöllecke J, Raidl S, Hüttl RF (2010) Defensive strategies of soil fungi to prevent grazing by Folsomia candida (Collembola). Pedobiologia 53:107–114
Bååth E, Söderström B (1980) Degradation of macromolecules by microfungi isolated from different podzolic soil horizons. Can J Bot 58:422–425
Berlemont R, Martiny AC (2015) Genomic potential for polysaccharide deconstruction in bacteria. Appl Environ Microbiol 81:1513–1519
Setälä H (1995) Growth of birch and pine seedlings in relation to grazing by soil fauna on ectomycorrhizal fungi. Ecology 76:1844–1851
Kaneda S, Kaneko N (2004) The feeding preference of a collembolan (Folsomia candida Willem) on ectomycorrhiza (Pisolithus tinctorius (Pers.)) varies with mycelial growth condition and vitality. Appl Soil Ecol 27:1–5
Thimm T, Hoffmann A, Borkott H, Munch JC, Tebbe CC (1998) The gut of the soil microarthropod Folsomia candida (Collembola) is a frequently changeable but selective habitat and a vector for microorganisms. Appl Environ Microbiol 64:2660–2669
Baldrian P, Voříšková J, Dobiášová P, Merhautová V, Lisá L, Valášková V (2011) Production of extracellular enzymes and degradation of biopolymers by saprotrophic microfungi from the upper layers of forest soil. Plant Soil 338:111–125
Brabcová V, Nováková M, Davidová A, Baldrian P (2016) Dead fungal mycelium in forest soil represents a decomposition hotspot and a habitat for a specific microbial community. New Phytol 210:1369–1381
Brabcová V, Štursová M, Baldrian P (2018) Nutrient content affects the turnover of fungal biomass in forest topsoil and the composition of associated microbial communities. Soil Biol Biochem 118:187–198
Drigo B, Anderson IC, Kannangara GS, Cairney JW, Johnson D (2012) Rapid incorporation of carbon from ectomycorrhizal mycelial necromass into soil fungal communities. Soil Biol Biochem 49:4–10
Nakamura N, Tanaka E, Tanaka C, Takeuchi-Kaneko Y (2018) Localization of helotialean fungi on ectomycorrhizae of Castanopsis cuspidata visualized by in situ hybridization. Mycorrhiza 28:17–28
Kerley SJ, Read DJ (1997) The biology of mycorrhiza in the Ericaceae XIX. Fungal mycelium as a nitrogen source for the ericoid mycorrhizal fungus Hymenoscyphus ericae (Read) Korf & Kernan and its host plants. New Phytol 136:691–701
Hodge A, Alexander IJ, Gooday GW (1995) Chitinolytic enzymes of pathogenic and ectomycorrhizal fungi. Mycol Res 99:935–941
Fernandez CW, Kennedy PG (2018) Melanization of mycorrhizal fungal necromass structures microbial decomposer communities. J Ecol 106:468–479
Baldrian P (2017) Microbial activity and the dynamics of ecosystem processes in forest soils. Curr Opin Microbiol 37:128–134
De Boer W, Folman LB, Summerbell RC, Boddy L (2005) Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev 29:795–811
Romaní AM, Fischer H, Mille-Lindblom C, Tranvik LJ (2006) Interactions of bacteria and fungi on decomposing litter: differential extracellular enzyme activities. Ecology 87:2559–2569
Halmschlager E, Kowalski T (2004) The mycobiota in nonmycorrhizal roots of healthy and declining oaks. Can J Bot 82:1446–1458
Fisk MC, Fahey TJ, Sobieraj JH, Staniec AC, Crist TO (2011) Rhizosphere disturbance influences fungal colonization and community development on dead fine roots. Plant Soil 341:279–293
Menkis A, Vasiliauskas R, Taylor AFS, Stenström E, Stenlid J, Finlay R (2006) Fungi in decayed roots of conifer seedlings in forest nurseries, afforested clear-cuts and abandoned farmland. Plant Pathol 55:117–129
Li A, Fahey TJ, Pawlowska TE, Fisk MC, Burtis J (2015) Fine root decomposition, nutrient mobilization and fungal communities in a pine forest ecosystem. Soil Biol Biochem 83:76–83
Hagerman SM, Jones MD, Bradfield GE, Gillespie M, Durall DM (1999) Effects of clear-cut logging on the diversity and persistence of ectomycorrhizae at a subalpine forest. Can J For Res 29:124–134
Erland S, Söderström B (1991) Effects of liming on ectomycorrhizal fungi infecting Pinus sylvestris L. III. Saprophytic growth and host plant infection at different pH values in unsterile humus. New Phytol 117:405–411
Cairney JWG, Burke RM (1994) Fungal enzymes degrading plant cell walls: their possible significance in the ectomycorrhizal symbiosis. Mycol Res 98:1345–1356
Lindahl BD, Finlay RD, Cairney J (2005) Enzymatic activities of mycelia in mycorrhizal fungal communities. In: Dighton J, White JF, Oudemans P (eds) The fungal community: its organization and role in the ecosystem. CRC Press, Boca Raton, pp 331–348
Giltrap NJ (1982) Production of polyphenol oxidases by ectomycorrhizal fungi with special reference to Lactarius spp. Trans Br Mycol Soc 78:75–81
Bödeker ITM, Nygren CMR, Taylor AFS, Olson A, Lindahl BD (2009) Class II peroxidase-encoding genes are present in a phylogenetically wide range of ectomycorrhizal fungi. ISME J 3:1387–1395
Rineau F, Roth D, Shah F, Smits M, Johansson T, Canbäck B, Olsen PB, Persson P, Grell MN, Lindquist E, Grigoriev IV (2012) The ectomycorrhizal fungus Paxillus involutus converts organic matter in plant litter using a trimmed brown-rot mechanism involving Fenton chemistry. Environ Microbiol 14:1477–1487
Nylund JE, Unestam T (1982) Structure and physiology of ectomycorrhizae I. The process of mycorrhiza formation in Norway spruce in vitro. New Phytol 91:63–79
Egger KN (1986) Substrate hydrolysis patterns of post-fire Ascomycetes (Pezizales). Mycologia 78:771–780
Egger KN (2006) The surprising diversity of ascomycetous mycorrhizas. New Phytol 170:421–423
Talbot JM, Allison SD, Treseder KK (2008) Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct Ecol 22:955–963
Lindahl BD, Tunlid A (2015) Ectomycorrhizal fungi–potential organic matter decomposers, yet not saprotrophs. New Phytol 205:1443–1447
Heinonsalo J, Sun H, Santalahti M, Bäcklund K, Hari P, Pumpanen J (2015) Evidences on the ability of mycorrhizal genus Piloderma to use organic nitrogen and deliver it to Scots pine. PLoS One 10:e0131561
Soil Classification Working Group (1998) The Canadian system of soil classification3rd edn. Agriculture and Agri-Food Canada Publication 1646, Ottawa
Martin KJ, Rygiewicz PT (2005) Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol 5:28
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, New York, pp 315–322
Kõljalg U, Larsson KH, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T (2005) UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166:1063–1068
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Keuskamp JA, Dingemans BJ, Lehtinen T, Sarneel JM, Hefting MM (2013) Tea Bag Index: a novel approach to collect uniform decomposition data across ecosystems. Methods Ecol Evol 4:1070–1075
Agerer R (2001) Exploration types of ectomycorrhizal mycelial systems. A proposal to classify mycorrhizal mycelial systems with respect to their ecologically important contact area with the substrate. Mycorrhiza 11:107–114
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118
Kernaghan G, Patriquin G (2015) Diversity and host preference of fungi co-inhabiting Cenococcum mycorrhizae. Fungal Ecol 17:84–95
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid Symp Ser 41:95–98
Rinaldi AC, Comandini O, Kuyper TW (2008) Ectomycorrhizal fungal diversity: separating the wheat from the chaff. Fungal Divers 33:1–45
Grünig CR, Queloz V, Sieber TN (2011) Structure of diversity in dark septate endophytes: from species to genes. In: Pirttila AM, Frank AC (eds) Endophytes of forest trees. Springer, Dordrecht, pp 3–30
Bödeker I, Lindahl BD, Olson Å, Clemmensen KE (2016) Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct Ecol 30:1967–1978
Nguyen NH, Song Z, Bates ST, Branco S, Tedersoo L, Menke J, Schilling JS, Kennedy PG (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248
Kernaghan G, Patriquin G (2011) Host associations between fungal root endophytes and boreal trees. Microb Ecol 62:460–473
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB, Simpson GL, Solymos P, Stevens MH, Wagner H (2013) Package ‘vegan’. Community ecology package, version 2(9)
Fernandez CW, Heckman K, Kolka R, Kennedy PG (2019) Melanin mitigates the accelerated decay of mycorrhizal necromass with peatland warming. Ecol Lett 22:498–505
LeFait A, Gailey J, Kernaghan G (2019) Fungal species selection during ectomycorrhizal grazing by Collembola. Symbiosis 78:87–95
Clericuzio M, Gilardoni G, Malagon O, Vidari G, Finz PV (2008) Sesquiterpenes of Lactarius and Russula (mushrooms): an update. Nat Prod Commun 3:951–974
Malagòn O, Porta A, Clericuzio M, Gilardoni G, Gozzini D, Vidari G (2014) Structures and biological significance of lactarane sesquiterpenes from the European mushroom Russula nobilis. Phytochemistry 107:126–134
Arocena JM, Glowa KR, Massicotte HB (2001) Calcium-rich hypha encrustations on Piloderma. Mycorrhiza 10:209–215
Whitney KD, Arnott HJ (1987) Calcium oxalate crystal morphology and development in Agaricus bisporus. Mycologia 79:180–187
Lawrence GB, Hazlett PW, Fernandez IJ, Ouimet R, Bailey SW, Shortle WC, Smith KT, Antidormi MR (2015) Declining acidic deposition begins reversal of forest-soil acidification in the northeastern US and eastern Canada. Environ Sci Technol 49:13103–13111
Schreiner T, Hildebrandt U, Bothe H, Marner FJ (1998) Chemical and biological characterization of corticrocin, a yellow pigment formed by the ectomycorrhizal fungus Piloderma croceum. Z Naturforsch C 53:4–8
Unestam T, Sun YP (1995) Extramatrical structures of hydrophobic and hydrophilic ectomycorrhizal fungi. Mycorrhiza 5:301–311
Lilleskov EA, Hobbie EA, Horton TR (2011) Conservation of ectomycorrhizal fungi: exploring the linkages between functional and taxonomic responses to anthropogenic N deposition. Fungal Ecol 4:174–183
Lenaers M, Reyns W, Czech J, Carleer R, Basak I, Deferme W, Krupinska P, Yildiz T, Saro S, Remans T, Vangronsveld J (2018) Links between heathland fungal biomass mineralization, melanization, and hydrophobicity. Microb Ecol 76:762–770
Pena R, Offermann C, Simon J, Naumann PS, Geßler A, Holst J, Dannenmann M, Mayer H, Kögel-Knabner I, Rennenberg H, Polle A (2010) Girdling affects ectomycorrhizal fungal (EMF) diversity and reveals functional differences in EMF community composition in a beech forest. Appl Environl Microbiol 76:1831–1841
Saravesi K, Aikio S, Wäli PR, Ruotsalainen AL, Kaukonen M, Huusko K, Suokas M, Brown SP, Jumpponen A, Tuomi J, Markkola A (2015) Moth outbreaks alter root-associated fungal communities in subarctic mountain birch forests. Microb Ecol 69:788–797
Hashimoto S, Suzuki M (2004) The impact of forest clear-cutting on soil temperature: a comparison between before and after cutting, and between clear-cut and control sites. J For Res 9:125–132
Solly EF, Schöning I, Boch S, Kandeler E, Marhan S, Michalzik B, Müller J, Zscheischler J, Trumbore SE, Schrumpf M (2014) Factors controlling decomposition rates of fine root litter in temperate forests and grasslands. Plant Soil 382:203–218
Lindahl BD, de Boer W, Finlay RD (2010) Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J 4:872–881
Yarwood SA, Myrold DD, Högberg MN (2006) Termination of belowground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microb Ecol 70:151–162
Schnecker J (2010) Ressourcenlimitierung von Abbauprozessen: Die Rolle von Pilzen. Thesis Magister der Naturwissenschaften (Mag Rer Nat). University of Vienna
Kohout P, Charvátová M, Štursová M, Mašínová T, Tomšovský M, Baldrian P (2018) Clearcutting alters decomposition processes and initiates complex restructuring of fungal communities in soil and tree roots. ISME J 12:692–703
Kolstad AL, Austrheim G, Solberg EJ, Venete AM, Woodin SJ, Speed JD (2018) Cervid exclusion alters boreal forest properties with little cascading impacts on soils. Ecosystems 21:1027–1041
Wang L, Otgonsuren B, Godbold DL (2017) Mycorrhizas and soil ecosystem function of co-existing woody vegetation islands at the alpine tree line. Plant Soil 411:467–481
Silver WL, Miya RK (2001) Global patterns in root decomposition: comparisons of climate and litter quality effects. Oecologia 129:407–419
Peterson RL, Massicotte HB, Melville LH (2004) Mycorrhizas: anatomy and cell biology. NRC Research Press, Ottawa
Downes GM, Alexander IJ, Cairney JWG (1992) A study of ageing of spruce [Picea sitchensis (Bong.) Carr.] ectomycorrhizas. I. Morphological and cellular changes in mycorrhizas formed by Tylospora fibrillosa (Burt.) Donk and Paxillus involutus (Batsch. ex Fr.) Fr. New Phytol 122:141–152
Philpott TJ, Barker JS, Prescott CE, Grayston SJ (2018) Limited effects of variable-retention harvesting on fungal communities decomposing fine roots in coastal temperate rainforests. Appl Environ Microbiol 84:1–16
Karst J, Erbilgin N, Pec GJ, Cigan PW, Najar A, Simard SW, Cahill JF (2015) Ectomycorrhizal fungi mediate indirect effects of a bark beetle outbreak on secondary chemistry and establishment of pine seedlings. New Phytol 208:904–914
Grelet GA, Ba R, Goeke DF, Houliston GJ, Taylor AF, Durall DM (2017) A plant growth-promoting symbiosis between Mycena galopus and Vaccinium corymbosum seedlings. Mycorrhiza 27:831–839
Halbwachs H, Dentinger BT, Detheridge AP, Karasch P, Griffith GW (2013) Hyphae of waxcap fungi colonise plant roots. Fungal Ecol 6:487–492
Tedersoo L, Pärtel K, Jairus T, Gates G, Põldmaa K, Tamm H (2009) Ascomycetes associated with ectomycorrhizas: molecular diversity and ecology with particular reference to the Helotiales. Environ Microbiol 11:3166–3178
Yamamoto S, Sato H, Tanabe AS, Hidaka A, Kadowaki K, Toju H (2014) Spatial segregation and aggregation of ectomycorrhizal and root-endophytic fungi in the seedlings of two Quercus species. PLoS One 9:e96363
Urban A, Puschenreiter M, Strauss J, Gorfer M (2008) Diversity and structure of ectomycorrhizal and co-associated fungal communities in a serpentine soil. Mycorrhiza 18:339–354
Sato H, Tanabe AS, Toju H (2015) Contrasting diversity and host association of ectomycorrhizal basidiomycetes versus root-associated ascomycetes in a dipterocarp rainforest. PLoS One 10:e0125550
Acknowledgments
We thank the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Province of Nova Scotia for funding, as well as the Nova Scotia Department of Natural Resources for accesses to field sites, the staff at the Mersey-Tobeatic Research Institute for their support, and Amanda Griffin for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Table S1
Identifications, GenBank accession numbers and functional guild designations of operational taxonomic units obtained from clone libraries. (XLSX 18 kb)
Rights and permissions
About this article
Cite this article
Gray, L., Kernaghan, G. Fungal Succession During the Decomposition of Ectomycorrhizal Fine Roots. Microb Ecol 79, 271–284 (2020). https://doi.org/10.1007/s00248-019-01418-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-019-01418-3