Abstract
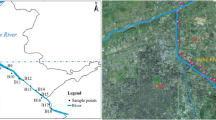
Pyrosequencing and metagenomic profiling were used to assess the phylogenetic and functional characteristics of microbial communities residing in sediments collected from the estuaries of Rivers Oujiang (OS) and Jiaojiang (JS) in the western region of the East China Sea. Another sediment sample was obtained from near the shore far from estuaries, used for contrast (CS). Characterization of estuary sediment bacterial communities showed that toxic chemicals potentially reduced the natural variability in microbial communities, while they increased the microbial metabolic enzymes and pathways. Polycyclic aromatic hydrocarbons (PAHs) and nitrobenzene were negatively correlated with the bacterial community variation. The dominant class in the sediments was Gammaproteobacteria. According to Kyoto Encyclopedia of Genes and Genomes (KEGG) enzyme profiles, dominant enzymes were found in estuarine sediments, which increased greatly, such as 2-oxoglutarate synthase, acetolactate synthase, inorganic diphosphatase, and aconitate hydratase. In KEGG pathway profiles, most of the pathways were also dominated by specific metabolism in these sediments and showed a marked increase, for instance alanine, aspartate, and glutamate metabolism, carbon fixation pathways in prokaryotes, and aminoacyl-tRNA biosynthesis. The estuarine sediment bacterial diversity varied with the polluted river water inputs. In the estuary receiving river water from the more seriously polluted River Oujiang, the sediment bacterial community function was more severely affected.




Similar content being viewed by others
References
Ahn IY, Kang YC, Choi JW (1995) The influence of industrial effluents on intertidal benthic communities in Panweol, Kyeonggi Bay (Yellow Sea) on the west coast of Korea. Mar Pollut Bull 30(3):200–206
Bairoch A, Boeckmann B, Ferro S, Gasteiger E (2004) SwissProt: juggling between evolution and stability. Brief Bioinform 5:39–55
Besaury L, Marty F, Buquet S, Mesnage V, Muyzer G, Quillet L (2012) Culture-dependent and independent studies of microbial diversity in highly copper-contaminated Chilean marine sediments. Microb Ecol 65(2):311–324
Buckley DH, Schmidt TM (2001) Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soils. FEMS Microbiol Ecol 35:105–112
Drury B, Rosi-Marshall E, Kelly JJ (2013) Wastewater treatment effluent reduces the abundance and diversity of benthic bacterial communities in urban and suburban rivers. Appl Environ Microbiol 79(6):1897–1905
Evans PR, Ward RM, Bone M, Leaky M (1998) Creation of temperate-climate intertidal mudflats: factors affecting colonization and use by benthic invertebrates and bird predators. Mar Pollut Bull 37(8–12):535–545
Ferrer M, Guazzaroni ME, Richter M, García-Salamanca A, Yarza P, Suárez-Suárez A, Solano J, Alcaide M, van Dillewijn P, Molina-HenaresMA L-CN, Al-Ramahi Y, Guerrero C, Acosta A, de Eugenio LI, Martínez V, Marques S, Rojo F, Santero E, Genilloud O, Pérez-Pérez J, Rosselló-Móra R, Ramos JL (2011) Taxonomic and functional metagenomic profiling of the microbial community in the anoxic sediment of a sub-saline shallow lake (Laguna de Carrizo, Central Spain). Microb Ecol 62(4):824–837
Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, Bateman A (2008) The Pfam protein families database. Nucleic Acids Res 36:281–288
Goñi-Urriza M, Capdepuy M, Raymond N, Quentin C, Caumette P (1999) Impact of an urban effluent on the bacterial community structure in the Arga River, Spain, with special reference to culturable Gram-negative rods. Can J Microbiol 45:826–832
Gray JP, Herwig RP (1996) Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol 62(11):4049–4059
Guazzaroni ME, Golyshin PN, Ferrer M (2010) Molecular methods to study complex microbial communities. In: Marco D (ed) Metagenomics: theory, methods and applications. Caister Academic Press, Norfolk, pp 55–77
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D (2009) InterPro: the integrative protein signature database. Nucleic Acids Res Database Issue 37:D224–D228
Huse SM, Huber JA, Morrison HG, Sogin ML, Welch DM (2007) Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8:149–155
Hyun S, Lee CH, Lee T, Choi JW (2007) Anthropogenic contributions to heavy metal distributions in the surface sediments of Masan Bay, Korea. Mar Pollut Bull 54(7):1059–1068
Jiang JH, Zhu LZ, Zhang M (2006) Concentration and sources of typical organic contaminants in seawater, sediment and organisms in Jiaojiang Bay. Environ Chem 25(5):546–549
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:480–484
Li M, Hong YG, Cao HL, Gu JD (2013) Community structures and distribution of anaerobic ammonium oxidizing and nirS-encoding nitrite-reducing bacteria in surface sediments of the South China Sea. Microb Ecol 66(2):281–296
Llobet-Brossa E, Russello-Mora R, Amann R (1998) Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol 64(7):2691–2696
Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J (2003) GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31:2187–2195
Needleman SB, Wunsch CD (1970) A general method applicable to the search for similarities in the amino acid sequences of two proteins. J Mol Biol 48:443–453
Noguchi H, Park J, Takagi T (2006) MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 34:5623–5630
Pang CM, Liu WT (2006) Biological filtration limits carbon availability and affects downstream biofilm formation and community structure. Appl Environ Microbiol 72:5702–5712
Richter M, Lombardot T, Kostadinov I, Kottmann R, Peplies J, Duhaime MB, Glöckner FO (2008) JCoast—a biologist-centric software tool for data mining and comparison of prokaryotic (meta)genomes. BMC Bioinf 9:1–9
Sonthiphand P, Hall MW, Neufeld JD (2014) Biogeography of anaerobic ammonia-oxidizing (anammox) bacteria. Front Microbiol 5:399
State Technology Supervision Bureau (2008) The specification for marine surveying (GB 12763—2008). Standards Press of China, Beijing
State Technology Supervision Bureau (2008) The specification for marine monitoring (GB 17378—2008). Standards Press of China, Beijing
Statistics Bureau of Zhejiang Province (2007) The development state and problems in ocean economy of Zhejiang Province. Conditions Strength China 5:63–64
Tang X, Gao G, Qin B, Zhu L, Chao J, Wang J, Yang G (2009) Characterization of bacterial communities associated with organic aggregates in a large, shallow, eutrophic freshwater lake (Lake Taihu, China). Microb Ecol 58:307–322
Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29:22–28
Urakawa H, Kita-Tsukamoto K, Ohwada K (1999) Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology 145:3305–3315
Vieites JM, Guazzaroni ME, Beloqui A, Golyshin PN, Ferrer M (2010) Molecular methods to study complex microbial communities. Methods Mol Biol 668:1–37
Vilchez R, Pozo C, Gómez MA, Rodelas B, González-López J (2007) Dominance of sphingomonads in a copper-exposed biofilm community for groundwater treatment. Microbiology 153:325–337
Wakelin SA, Colloff MJ, Kookana RS (2008) Effect of wastewater treatment plant effluent on microbial function and community structure in the sediment of a freshwater stream with variable seasonal flow. Appl Environ Microbiol 74:2659–2668
Wang Y, Sheng HF, He Y, Wu JY, Jiang YX, Tam NFY, Zhou HW (2012) Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl Environ Microbiol 78(23):8264–8271
Zhang S, Zhang C, Tian X, Wang FA (2010) The study of diversities of marine microbes in China. Bull Chin Acad Sci 24(4):228–234
Zhang W, Ki J, Qian P (2008) Microbial diversity in polluted harbor sediments I: bacterial community assessment based on four clone libraries of 16S rDNA. Estuar Coast Shelf Sci 76(3):668–681
Zhao YQ, Zeng JN, Gao AG, Chen QZ, Liao YB, Shou L (2009) Community pattern and diversity of macrozoobenthos in an intertidal flat, Jiaojiang Estuary. Biodivers Sci 17(3):303–309
Acknowledgments
This work is supported by Open Project Program of the Key Laboratory of Tropical Marine Bio-resources and Ecology, SCSIO, CAS (No. LMB141001). This work is supported by MEL Visiting Fellowship of State Key Laboratory of Marine Environmental Science, Xiamen University (No. MELRS1507). This work is supported by the Science and Technology Program of Wenzhou, China (No. S20140004). This work is supported by the Science and Technology Innovation Project of Water Pollution Control and Treatment of Wenzhou City (No. S20150001).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 964 kb)
Rights and permissions
About this article
Cite this article
Lu, XM., Chen, C. & Zheng, TL. Metagenomic Insights into Effects of Chemical Pollutants on Microbial Community Composition and Function in Estuarine Sediments Receiving Polluted River Water. Microb Ecol 73, 791–800 (2017). https://doi.org/10.1007/s00248-016-0868-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-016-0868-8