Abstract
Rock varnish is a thin layer of Fe and Mn oxyhydroxides with embedded clay minerals that contain an increased Mn/Fe ratio compared to that of the Earth’s crust. Even if the study of rock varnish has important implications in several fields, the composition of epilithic bacterial communities and the distribution of taxa on varnish surfaces are still not wholly described. The aim of this study was (i) to identify the bacterial taxa which show the greatest variation between varnish and non-varnish environments, collected from the same rock, and (ii) to describe the morphology of epilithic communities through scanning electron microscopy (SEM). Triplicate samples of rock surfaces with varnish and triplicate samples without varnish were collected from five sites in Matsch Valley (South Tyrol, Italy). The V4 region of 16S rRNA gene was analyzed by Illumina sequencing. Fifty-five ubiquitous taxa have been examined to assess variation between varnish and non-varnish. Cyanobacteria, Chloroflexi, Proteobacteria along with minor taxa such as Solirubrobacterales, Conexibaxter, and Rhodopila showed significant variations of abundance, diversity, or both responding to the ecology (presence/absence of varnish). Other taxa, such as the genus Edaphobacter, showed a more marked spatial variation responding to the sampling site. SEM images showed a multitude of bacterial morphologies and structures involved in the process of attachment and creation of a suitable environment for growth. The features emerging from this analysis suggest that the highly oxidative Fe and Mn-rich varnish environment favors anoxigenic autotrophy and establishment of highly specialized bacteria.



Similar content being viewed by others
References
Dorn R (2007) Rock varnish. Geochemical sediments and landscapes. Wiley Online Library, pp 246–297
Liu T, Broecker WS (2000) How fast does rock varnish grow? Geology 28:183–186
Engel CG, Sharp RP (1958) Chemical data on desert varnish. Geol Soc Am Bull 69:487–518
Krinsley D, Dorn RI, DiGregorio B (2009) Astrobiological implications of rock varnish in Tibet. Astrobiology 9:551–562
Northup DE, Snider JR, Spilde MN et al (2010) Diversity of rock varnish bacterial communities from Black Canyon, New Mexico. J Geophys Res-Biogeo 115:1–19
Liu T, Broecker WS, Stein M (2013) Rock varnish evidence for a younger Dryas wet period in the Dead Sea Basin. Geophys Res Lett 40:2229–2235
Eggleston CM, Parkinson BA (2012) Natural solar cells on Earth and Mars. Goldshmidt Conf Abstr 1678
Hooke RL, Yang H-Y, Weiblen PW (1969) Desert varnish: an electron probe study. J Geol 77:275–288
Allen CC, Probst LW, Flood BE et al (2004) Meridiani Planum hematite deposit and the search for evidence of life on Mars—Iron mineralization of microorganisms in rock varnish. Icarus 171:20–30
Perry RS, Kolb VM (2004) Biological and organic constituents of desert varnish: review and new hypotheses. Opt Sci Technol SPIE’s 48th Annu Meet pp 202–217
Mergelov NS, Goryachkin SV, Shorkunov IG et al (2012) Endolithic pedogenesis and rock varnish on massive crystalline rocks in East Antarctica. Eurasian Soil Sci 45:901–917
Weber KA, Achenbach LA, Coates JD (2006) Microbes pumping iron: anaerobic microbial iron oxidation and reduction. Nat Rev Microbiol 4:752–764
Tebo BM, Johnson HA, McCarthy JK, Templeton AS (2005) Geomicrobiology of manganese(II) oxidation. Trends Microbiol 13:421–428
Learman DR, Voelker BM, Vazquez-Rodriguez AI, Hansel CM (2011) Formation of manganese oxides by bacterially generated superoxide. Nat Geosci 4:95–98
Krumbein WE, Jens K (1981) Biogenic rock varnishes of the Negev Desert (Israel) an ecological study of iron and manganese transformation by cyanobacteria and fungi. Oecologia 50:25–38
Kuhlman KR, Fusco WG, La Duc MT et al (2006) Diversity of microorganisms within rock varnish in the Whipple Mountains, California. Appl Environ Microbiol 72:1708–1715
Krinsley DH, Dorn RI, DiGregorio BE et al (2012) Rock varnish in New York: an accelerated snapshot of accretionary processes. Geomorphology 138:339–351
Penna D, Engel M, Mao L et al (2014) Tracer-based analysis of spatial and temporal variation of water sources in a glacierized catchment. Hydrol Earth Syst Sci Discuss 11:4879–4924
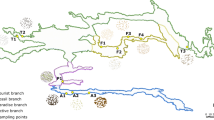
Habler G, Thöni M, Grasemann B (2009) Cretaceous metamorphism in the Austroalpine Matsch Unit (Eastern Alps): the interrelation between deformation and chemical equilibration processes. Mineral Petrol 97:149–171
Knoll C, Kerschner H (2010) A glacier inventory for South Tyrol, Italy, based on airborne laser-scanner data. Ann Glaciol 50:46–52
Feinstein LM, Sul WJ, Blackwood CB (2009) Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl Environ Microbiol 75:5428–5433
Lane DS, Stackebrandt E, Goodfellow M (1990) 16S and 23S rRNA sequencing. In: Stackebrandt E and Goodfellow M (ed) Nucleic Acid Tech Bact Syst NY, John Wiley and Sons, pp 115–175
Caporaso JG, Lauber CL, Walters WA et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522
Echlin P (2009) Handbook of sample preparation for scanning electron microscopy and X-ray microanalysis. Springer
Aronesty E (2013) Comparison of sequencing utility programs. TOBIOIJ 7:1–8
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
R Core Team (2012). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/
Cole JR, Wang Q, Cardenas E et al (2009) The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145
Schloss PD, Westcott SL, Ryabin T et al (2009) Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Quast C, Pruesse E, Yilmaz P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596
Price MN, Dehal PS, Arkin AP (2010) FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490
Hamady M, Lozupone C, Knight R (2010) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27
Parchert KJ, Spilde MN, Porras-Alfaro A et al (2012) Fungal communities associated with rock varnish in Black Canyon, New Mexico: casual inhabitants or essential partners? Geomicrobiol J 29:752–766
Marnocha CL, Dixon JC (2013) Bacterially facilitated rock-coating formation as a component of the geochemical budget in cold climates: an example from Kärkevagge, Swedish Lapland. Geomorphology 218:41–51
Claesson MJ, Wang Q, O’Sullivan O et al (2010) Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200
Yabe S, Aiba Y, Sakai Y et al (2011) Thermogemmatispora onikobensis gen. nov., sp. nov. and Thermogemmatispora foliorum sp. nov., isolated from fallen leaves on geothermal soils, and description of Thermogemmatisporaceae fam. nov. and Thermogemmatisporales. Int J Syst Evol Microbiol 61:903–910
Jones RT, Robeson MS, Lauber CL et al (2009) A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3:442–453
Catão ECP, Lopes FAC, Araújo JF et al (2014) Soil Acidobacterial 16S rRNA gene sequences reveal subgroup level differences between Savanna-Like Cerrado and Atlantic Forest Brazilian biomes. Int J Microbiol 2014:156341
Kuhlman KR, Venkat P, La Duc MT et al (2008) Evidence of a microbial community associated with rock varnish at Yungay, Atacama Desert, Chile. J Geophys Res 113:G04022
Gupta RS, Chander P, George S (2013) Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexi class. nov. into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochlorida. Antonie Van Leeuwenhoek 103:99–119
Benning MM, Meyer TE, Holden HM (1996) Molecular structure of a high potential cytochrome c2 isolated from Rhodopila globiformis. Arch Biochem Biophys 333:338–348
Taylor-George S, Palmer F, Staley JT et al (1983) Fungi and bacteria involved in desert varnish formation. Microb Ecol 9:227–245
Ehrlich HL, Newman DK (2008) Geomicrobiology, 5th edn. CRC Press, Boca Raton, 656 p
Jiao Y, Kappler A, Croal LR, Johnson DB (2005) Isolation and characterization of a genetically tractable photoautotrophic Fe(II)-oxidizing bacterium, Rhodopseudomonas palustris strain TIE-1. Appl Environ Microbiol 71(8):4487–4496
Caccavo FJ, Das A (2002) Adhesion of dissimilatory Fe(III)-reducing bacteria to Fe(III) minerals. Geomicrobiol J 19:161–177
Roberts JA (2004) Inhibition and enhancement of microbial surface colonization: the role of silicate composition. Chem Geol 212:313–327
Gruszczynski M, Kowalski B, Soltysik R, Hercman H (2004) Tectonic origin of the unique Holocene travertine from the Holy Cross Mts.: microbially and abiologically mediated calcium carbonate, and manganese oxide precipitation. Acta Geol Pol 54:61–76
Wang X, Zeng L, Wiens M et al (2011) Evidence for a biogenic, microorganismal origin of rock varnish from the Gangdese Belt of Tibet. Micron 42:401–411
Ivarsson M, Holmström S (2012) The use of ESEM in geobiology, scanning electron microscopy, Dr Viacheslav Kazmiruk (Ed), ISBN: 978-953-51-0092-8, InTech
Acknowledgments
We acknowledge the help of Kjell Jansson for his suggestions for SEM analysis, Boyke Bunk for bioinformatics, and Anders Andersson for his help in statistics. The present study was supported by the Erich-Ritter and the Herzog-Sellenberg Foundations within the Stifterverband für die Deutsche Wissenschaft within the EMERGE project (CUP I41J11000490007), and the grants from Liljevalchs fund, Stockholm University. English quality was edited by Dr. James Haile and Hilderd Crill.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. SM1
Shannon (upper row) and Chao (lower row) diversity indices for Cyanobacteria, Solirubrobacterales and Conexibacter, and table with diversity indices values for each of the sites. (PPT 150 kb)
Fig. SM2
Bacterial community profile based on Illumina sequencing (on average 354494 reads per sample). Major (> 99.9 % of the reads) phyla, classes and orders are depicted. The table in the lower right quarter of the picture is a list of the other taxa divided according to their occurrence in one of the two environment or in both; the number of samples in which the taxa occur are given in the parenthesis. (PPT 385 kb)
Fig. SM3
Interactions between microbes and minerals: (A) structure analogous to figure 3G; with stone surface corrosion by filamentous bacteria from site 3 varnish; (B) filamentous forms colonizing a microfracture from non-varnish of site 4; (C) unknown structures from varnish of site 5; (D) unknown blood-cell like shapes from non-varnish of site 2; (E) cave buds of unknown origin from non-varnish sample of site 3; (F) putative rock-dwelling microcolonial fungi, from varnish of site 5; (G) analogous structure as in (F) plus enlargements of the biological structure (H-I) from varnish of site 3. (PPT 2333 kb)
Fig. SM4
Landscape pictures, extracellular structure and dividing cells: (A) micrometre thick coating on mineral surface from varnish of site 4; (B) landscape aspect of non-varnish samples of site 4; (C) filamentous form from varnish of site 3; (D) tetra-cocci with external slime from varnish of site 2; (E) coccal shape with external biofilms from varnish of site 3 and (F) varnish site 2; (G) coccal shapes adhering to the mineral surface, zoom from figure 2c; (H) reproducing bacteria from varnish of site 3 and (I) non-varnish of site 2. (PPT 2050 kb)
Rights and permissions
About this article
Cite this article
Esposito, A., Ahmed, E., Ciccazzo, S. et al. Comparison of Rock Varnish Bacterial Communities with Surrounding Non-Varnished Rock Surfaces: Taxon-Specific Analysis and Morphological Description. Microb Ecol 70, 741–750 (2015). https://doi.org/10.1007/s00248-015-0617-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-015-0617-4