Abstract
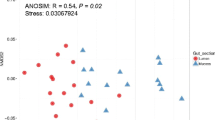
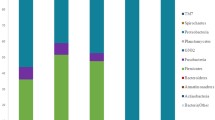
Dominant bacterial microbiota of the gut of juvenile farmed Atlantic salmon was investigated using a combination of molecular approaches. Bacterial community composition from the stomach, the pyloric caeca, and the intestine was assessed by extracting DNA directly from each gut compartment. Temporal temperature gradient gel electrophoresis (TTGE) analysis of 16S ribosomal DNA (rDNA) amplicons showed very similar bacterial compositions throughout the digestive tract. Band sequencing revealed a narrow diversity of species with a dominance of Pseudomonas in the three compartments. However, cloning revealed more diversity among the Pseudomonas sequences. To confirm these results, we analyzed the bacterial community by amplifying the variable 16S–23S rDNA intergenic spacer region (ITS). Similar ITS profiles were observed among gastrointestinal compartments of salmon, confirming the TTGE results. Moreover, the dominant ITS band at 650 bp, identified as Pseudomonas, was observed in the ITS profile from fish collected in two seasons (July 2003 and 2004). In contrast, aerobic culture analysis revealed Shewanella spp. as the most prevalent isolate. This discrepancy was resolved by evaluating 16S rDNA and ITS polymerase chain reaction amplification efficiency from both Shewanella and Pseudomonas isolates. Very similar efficiencies were observed in the two bacteria. Hence, this discrepancy may be explained by preferential cultivation of Shewanella spp. under the experimental conditions. Also, we included analyses of pelleted feed and the water influent to explore environmental influences on the bacterial composition of the gut microbiota. Overall, these results indicate a homogeneous composition of the bacterial community composition along the gastrointestinal tract of reared juvenile salmon. This community is mainly composed of Pseudomonas spp., which could be derived from water influent and may be selectively associated with salmon in this hatchery.




Similar content being viewed by others
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in-situ detection of individual microbial-cells without cultivation. Microbiol Rev 59:143–169
Andlid T, Vazquez-Juarez R, Gustafsson L (1998) Yeasts isolated from the intestine of rainbow trout adhere to and grow in intestinal mucus. Mol Mar Biol Biotech 7:115–126
Austin B (2006) The bacterial microflora of fish, revised. ScientificWorldJournal 6:931–945
Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101:15718–15723
Bates J, Mittg E, Kuhlman J, Baden K, Cheesman S, Guindulain T (2006) Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol 297:374–386
Bergot P, Solari A, Luquet P (1975) Comparaison des surfaces absorbantes des caeca pyloriques et de l’intestin chez la truite arc-en-ciel (Salmo gairdneri Rich). Ann Hydrobiol 6:27–43
Buddington RK, Chen JW, Diamond J (1987) Genetic and phenotypic adaptation of intestinal nutrient transport to diet in fish. J Physiol 393:261–281
Buddington RK, Diamond JM (1987) Pyloric ceca of fish: a “new” absorptive organ. Am J Physiol 252:G65–G76
Buddington RK, Krogdahl A, BakkeMcKellep AM (1997) The intestines of carnivorous fish: structure and functions and the relations with diet. Acta Physiol Scand 161:67–80
Cahill MM (1990) Bacterial-flora of fish—a review. Microb Ecol 19:21–41
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM (2005) The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 33:D294–D296
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35:D169–D172
Espejo RT, Escanilla D (1993) Detection of HIV1 DNA by a simple procedure of polymerase chain reaction, using “primer-dimer” formation as an internal control of amplification. Res Virol 144:243–246
Espejo RT, Feijoo CG, Romero J, Vasquez M (1998) PAGE analysis of the heteroduplexes formed between PCR-amplified 16S rRNA genes: estimation of sequence similarity and rDNA complexity. Microbiology 144(Pt 6):1611–1617
Espejo RT, Romero J (1997) Bacterial community in copper sulfide ores inoculated and leached with solution from a commercial-scale copper leaching plant. Appl Environ Microbiol 63:1344–1348
Gordon JI (2005) A genomic view of our symbiosis with members of the gut microbiota. J Pediatr Gastroenterol Nutr 40(Suppl 1):S28
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Griffiths S, Melville K, Cook M, Vincent S, Pierre M, Lanteigne C (2001) Profiling of bacterial species associated with haddock larviculture by PCR amplification of 16S rDNA and denaturing gradient gel electrophoresis. J Aquat Anim Health 13:355–363
Hansen GH, Olafsen JA (1999) Bacterial interactions in early life stages of marine cold water fish. Microb Ecol 38:1–26
Holben WE, Williams P, Gilbert MA, Saarinen M, Sarkilahti LK, Apajalahti JH (2002) Phylogenetic analysis of intestinal microflora indicates a novel Mycoplasma phylotype in farmed and wild salmon. Microb Ecol 44:175–185
Hovda MB, Lunestad BT, Fontanillas R, Rosnes JT (2007) Molecular characterisation of the intestinal microbiota of farmed Atlantic salmon (Salmo salar L.). Aquaculture 272:581–588
Jensen MA, Webster JA, Straus N (1993) Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol 59:945–952
Jensen S, Øvreås L, Bergh O, Torsvik V (2004) Phylogenetic analysis of bacterial communities associated with larvae of the Atlantic halibut propose succession from a uniform normal flora. Syst Appl Microbiol 27:728–736
Kim DH, Brunt J, Austin B (2007) Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhynchus mykiss). J Appl Microbiol 102:1654–1664
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651
Magne F, Abely M, Boyer F, Morville P, Pochart P, Suau A (2006) Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol 57:128–138
McCracken VJ, Simpson JM, Mackle RI, Gaskins HR (2001) Molecular ecological analysis of dietary and antibiotic-induced alterations of the mouse intestinal microbiota. J Nutr 131:1862–1870
Moreno C, Romero J, Espejo RT (2002) Polymorphism in repeated 16S rRNA genes is a common property of type strains and environmental isolates of the genus Vibrio. Microbiology 148:1233–1239
Muyzer G, Dewaal EC, Uitterlinden AG (1993) Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16S ribosomal-RNA. Appl Environ Microbiol 59:695–700
Olafsen JA (2001) Interactions between fish larvae and bacteria in marine aquaculture. Aquaculture 200:223–247
Pond MJ, Stone DM, Alderman DJ (2006) Comparison of conventional and molecular techniques to investigate the intestinal microflora of rainbow trout (Oncorhynchus mykiss). Aquaculture 261:194–203
Ransom D, Lannan C, Rohovec J, Fryer J (1984) Comparison of histopathology caused by Vibrio anguillarum and Vibrio ordalii in three species of Pacific salmon. J Fish Dis 7:107–115
Rawls JF, Mahowald MA, Ley RE, Gordon JI (2006) Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433
Rawls JF, Samuel BS, Gordon JI (2004) Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A 101:4596–4601
Rawls JF, Mahowald MA, Goodman AL, Trent CM, Gordon JI (2007) In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc Natl Acad Sci U S A 104:7622–7627
Ringø E, Birkbeck T (1999) Intestinal microflora of fish larvae and fry. Aquac Res 30:73–93
Ringø E, Olsen RE, Mayhew TM, Myklebust R (2003) Electron microscopy of the intestinal microflora of fish. Aquaculture 227:395–415
Ringø E, Sperstad S, Myklebust R, Refstie S, Krogdahl A (2006) Characterisation of the microbiota associated with intestine of Atlantic cod (Gadus morhua L.): the effect of fish meal, standard soybean meal and a bioprocessed soybean meal. Aquaculture 261:829–841
Ringø E, Strom E, Tabachek J (1995) Intestinal microflora of salmonids: a review. Aquac Res 26:773–789
Romero J, Espejo R (2001) The prevalence of noncultivable bacteria in oysters (Tiostrea chilensis, Philippi, 1845). J Shellfish Res 20:1235–1240
Romero J, Garcia-Varela M, Laclette JP, Espejo RT (2002) Bacterial 16S rRNA gene analysis revealed that bacteria related to Arcobacter spp. constitute an abundant and common component of the oyster microbiota (Tiostrea chilensis). Microb Ecol 44:365–371
Romero J, Navarrete P (2006) 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch). Microb Ecol 51:422–430
Rønnestad I, Rojas-Garcia CR, Skadal J (2000) Retrograde peristalsis; a possible mechanism for filling the pyloric caeca? J Fish Biol 56:216–218
Rust M (2002) Nutritional physiology. In: Halver J, Hardy R (eds) Fish nutrition. Academic, San Diego, pp 368–446
Sonnenburg JL, Angenent LT, Gordon JI (2004) Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol 5:569–573
Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, Dore J (1999) Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65:4799–4807
Suyehiro Y (1942) A study on the digestive system and feeding habits of fish. Jpn J Zool 10:1–303
vandePeer Y, DeWachter R (1997) Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci 13:227–230
Verschuere L, Rombaut G, Sorgeloos P, Verstraete W (2000) Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev 64:655–671
von Wintzingerode F, Göbel U, Stackebrandt E (1997) Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev 21:213–229
Acknowledgments
This project was supported by a grant (FONDECYT No. 1061121) from CONICYT-Chile. P. Navarrete acknowledges a scholarship from CONICYT-Chile and Dr. Stekel a fellowship from INTA-Nestlé. Partial support was derived from an INNOVA CORFO grant (05CT6PPT-09) and FONDECYT 1080480.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Navarrete, P., Espejo, R.T. & Romero, J. Molecular Analysis of Microbiota Along the Digestive Tract of Juvenile Atlantic Salmon (Salmo salar L.). Microb Ecol 57, 550–561 (2009). https://doi.org/10.1007/s00248-008-9448-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-008-9448-x