Abstract
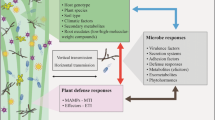
Systemic acquired resistance (SAR) is an inducible systemic plant defense against a broad spectrum of plant pathogens, with the potential to secrete antimicrobial compounds into the soil. However, its impact on rhizosphere bacteria is not known. In this study, we examined fingerprints of bacterial communities in the rhizosphere of the model plant Arabidopsis thaliana to determine the effect of SAR on bacterial community structure and diversity. We compared Arabidopsis mutants that are constitutive and non-inducible for SAR and verified SAR activation by measuring pathogenesis-related protein activity via a β-glucoronidase (GUS) reporter construct driven by the β-1-3 glucanase promoter. We used terminal restriction fragment length polymorphism (T-RFLP) analysis of MspI- and HaeIII-digested 16S rDNA to estimate bacterial rhizosphere community diversity, with Lactobacillus sp. added as internal controls. T-RFLP analysis showed a clear rhizosphere effect on community structure, and diversity analysis of both rhizosphere and bulk soil operational taxonomic units (as defined by terminal restriction fragments) using richness, Shannon–Weiner, and Simpson’s diversity indices and evenness confirmed that the presence of Arabidopsis roots significantly altered bacterial communities. This effect of altered soil microbial community structure by plants was also seen upon multivariate cluster analysis of the terminal restriction fragments. We also found visible differences in the rhizosphere community fingerprints of different Arabidopsis SAR mutants; however, there was no clear decrease of rhizosphere diversity because of constitutive SAR expression. Our study suggests that SAR can alter rhizosphere bacterial communities, opening the door to further understanding and application of inducible plant defense as a driving force in structuring soil bacterial assemblages.



Similar content being viewed by others
References
Bankhead SB, Landa BB, Lutton E, Weller DM, McSpadden Gardener BB (2004) Minimal changes in rhizobacterial population structure following root colonization by wild type and transgenic biocontrol strains. FEMS Microbiol Ecol 49:307–318
Baudoin E, Benizri E, Guckert A (2003) Impact of artificial root exudates on the bacterial community structure in bulk soil and maize rhizosphere. Soil Biol Biochem 35:1183–1192
Becker S, Boger P, Oehlmann R, Ernst A (2000) PCR bias in ecological analysis: a case study for quantitative Taq nuclease assays in analyses of microbial communities. Appl Environ Microbiol 66:4945–4953
Bent SJ, Pierson JD, Forney LJ, Danovaro R, Luna GM, Dell’Anno A, Pietrangeli B (2007) Measuring species richness based on microbial community fingerprints: the emperor has no clothes. Appl Environ Microbiol 73:2399–2401
Blackwood CB, Marsh T, Kim S-H, Eldor P (2003) Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl Environ Microbiol 69:926–932
Bowling SA, Guo A, Cao H, Gordon SA, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6:1845–1857
Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13:1499–1510
Burketova L, Stillerova K, Feltlova M (2003) Immunohistological localization of chitinase and b-1,3-glucanase in rhizomania-diseased and benzothiadiazole treated sugar beet roots. Physiol Mol Plant Pathol 63:47–54
Cameron RK, Paiva NL, Lamb CJ, Dixon RA (1999) Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance (SAR) response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiol Mol Plant Pathol 55:121–130
Cao H, Bowling SA, Gordon SA, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6:1583–1592
Conrath U, Zhixiang C, Ricigliano JR, Klessig DF (1995) Two inducers of plant defense responses, 2,6-dichloroisonicotinic acid and salicylic acid, inhibit catalase activity in tobacco. Proc Natl Acad Sci USA 92:7143–7147
Curl EA, Truelove B (1986) The Rhizosphere. Springer, Berlin
Devoto A, Turner JG (2003) Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann Bot 92:329–337
Dong X (2004) NPR1, all things considered. Curr Opin Plant Biol 7:547–552
Dunbar JM, Ticknor LO, Kuske CR (2000) Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl Environ Microbiol 66:2943–2950
Dunbar JM, Ticknor LO, Kuske CR (2001) Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl Environ Microbiol 67:190–197
Dunfield KE, Germida JJ (2003) Seasonal changes in the rhizosphere microbial communities associated with field-grown genetically modified canola (Brassica napus). Appl Environ Microbiol 69:7310–7318
Egert M, Friedrich MW (2003) Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl Environ Microbiol 69:2555–2562
Fierer N, Schimel JP, Holden PA (2003) Influence of drying-rewetting frequency on soil bacterial community structure. Microb Ecol 45:63–71
Friedrich L, Lawton K, Ruess W, Masner P, Specker N, Gut Rella M, Meier B, Dincher S, Staub T, Uknes S, Metraux JP, Kessman H, Ryals J (1996) A benzothiadiaxole derivative induces systemic acquired resistance in tobacco. Plant J 10:61–70
Fromin N, Achouak W, Thiery JM, Heulin T (2001) The genotypic diversity of Pseudomonas brassicacearum populations isolated from roots of Arabidopsis thaliana: influence of plant genotype. FEMS Microbiol Ecol 37:21–29
Gessler C, Kuc J (1982) Induction of resistance to Fusarium wilt in cucumber by root and foliar pathogens. Phytopathology 72:1439–1441
Grant M, Lamb CJ (2006) Systemic immunity. Curr Opin Plant Biol 9:414–420
Griffiths BS, Ritz K, Ebblewhite N, Dobson G (1999) Soil microbial community structure: effects of substrate loading rates. Soil Biol Biochem 31:145–153
Hackl E, Zechmeister-Boltenstern S, Bodrossy L, Sessitsch A (2004) Comparison of diversities and compositions of bacterial populations inhabiting natural forest soils. Appl Environ Microbiol 70:5057–5065
Heidel AJ, Clarke J, Antonovics J, Dong X (2004) Fitness costs of mutations affecting the systemic acquired resistance pathway in Arabidopsis thaliana. Genetics 168:2197–2206
Heil M (1999) Systemic acquired resistance: available information and open ecological questions. J Ecol 87:341–346
Heil M, Baldwin IT (2002) Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci 7:61–67
Heil M, Hilpert A, Kaiser W, Linsenmair KE (2000) Reduced growth and seed set following chemical induction of pathogen defense: does systemic acquired resistance (SAR) incur allocation costs? J Ecol 88:645–654
Kerkhof L, Santoro M, Garland J (2000) Response of soybean rhizosphere communities to human hygiene water addition as determined by community level physiological profiling (CLPP) and terminal restriction fragment length polymorphism (TRFLP) analysis. FEMS Microbiol Lett 184:95–101
Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD (2004) Molecular Microbial Ecology Manual, 2nd edn., vols. 1 and 2. Kluwer, Dordrecht, The Netherlands
Kuske CR, Ticknor LO, Miller ME, Dunbar JM, Davis JA, Barns SM, Belnap J (2002) Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Appl Environ Microbiol 68:1854–1863
Lukow T, Dunfield PF, Liesack W (2000) Use of the T-RFLP technique to assess spatial and temporal changes in the bacterial community structure within an agricultural soil planted with transgenic and non-transgenic potato plants. FEMS Microbiol Ecol 32:241–247
Malamy J, Hennig J, Klessig DF (1992) Temperature-dependent induction of Salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell 4:359–366
Marilley L, Vogt G, Blanc M, Aragno M (1998) Bacterial diversity in the bulk soil and rhizosphere fractions of Lolium perenne and Trifolium repens as revealed by PCR restriction analysis of 16S rDNA. Plant Soil 198:219–224
Marschner P, Yang C-H, Lieberei R, Crowley DE (2001) Soil and plant specific effects on bacterial community composition in the rhizosphere. Soil Biol Biochem 33:1437–1445
Martin AP (2002) Phylogenetic apporaches for describing and comparing the diversity of microbial communities. Appl Environ Microbiol 68:3673–3682
Moeseneder MM, Arrieta JM, Muyzer G, Winter C, Herndl G (1999) Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol 65:3518–3525
O’Donnell AG, Seasman M, Macrae A, Waite I, Davies JT (2001) Plants and fertilisers as drivers of change in microbial community structure and function in soils. Plant Soil 232:135–145
Pieterse CMJ, van Loon LC (1999) Salicylic acid-independent plant defence pathways. Trends Plant Sci 4:52–58
Pieterse CMJ, van Wees SCM, van Pelt JA, Knoester M, Laan R, Gerrits H, Weisbeek PJ, van Loon LC (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580
Rees GN, Baldwin DS, Watson GO, Perryman S, Nielsen DL (2004) Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Antonie van Leeuwenhoek 86:339–347
Ryals J, Uknes S, Ward E (1994) Systemic acquired resistance. Plant Physiol 104:1109–1112
Santamaria M, Thomson CJ, Read ND, Loake GJ (2001) The promoter of a basic PR1-like gene, AtPRB1, from Arabidopsis establishes an organ-specific expression pattern and responsiveness to ethylene and methyl jasmonate. Plant Mol Biol 47:641–652
Sauerborn J, Buschmann K, Ghiasvand Ghiasi K, Kogel K-H (2002) Benzothiadiazole activates resistance in sunflower (Helianthus annuus) to the root-parasitic weed Orobanche cumana. Phytopathology 92:59–64
Schutter M, Dick R (2001) Shifts in substrate utilization potential and structure of soil microbial communities in response to carbon substrates. Soil Biol Biochem 33:1481–1491
Sessitsch A, Reiter B, Berg G (2004) Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities. Can J Microbiol 50:239–249
Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E (2001) Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microbiol 67:4215–4224
Shah J (2003) The salicylic acid loop in plant defense. Curr Opin Plant Biol 6:365–371
Sliwinski MK, Goodman RM (2004) Comparison of crenarchaeal consortia inhabiting the rhizosphere of diverse terrestrial plants with those in bulk soil in native environments. Appl Environ Microbiol 70:1821–1826
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer, H, Berg G. (2001) Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol 67:4742–4751
Sticher L, Mauch-Mani B, Metraux JP (1997) Systemic acquired resistance. Ann Rev Phytopathol 35:235–270
Stokes TL, Richards EJ (2002) Induced instability of two Arabidopsis constitutive pathogen-response alleles. Proc Natl Acad Sci USA 99:7792–7796
Suzuki MT, Giovannoni SJ (1996) Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol 62:625–630
Tahiri-Alaoui A, Dumas-Gaudot E, Gianinazzi S, Antoniw J (1993) Expression of the PR-bi gene in roots of two Nicotiana species and their amphidipioid hybrid infected with virulent and avirulent races of Chalara elegans. Plant Pathol 42:728–736
Tebbe CC, Vahjen W (1993) Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol 59:2657–2665
van Loon LC, van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
Walker T, Pal Bais H, Grotewold E, Vivanco JM (2003) Root exudation and rhizosphere biology. Plant Physiol 132:44–51
Walker T, Pal Bais H, Halligan KM, Stermitz FR, Vivanco JM (2003) Metabolic profiling of root exudates of Arabidopsis thaliana. J Agric Food Chem 51:2548–2554
Acknowledgments
The seeds used in this project were kindly provided by Dr. X. Dong (Duke University). Mark Wilson (Humboldt State Univ.) gave advice on preparing T-RFLP samples. Alan Raetz (CSUC) wrote PERL programs that helped with initial data analysis. Larry Hanne and Nancy Carter (CSUC) gave helpful suggestions. The ABI 310 genetic analyzer was funded by NSF CCLI #DUE-0126618 to GW. GW and KB received funds for this project from a CSU Biotechnology consortium (CSUPERB) faculty seed grant, and JH received funding from the Sigma Xi Grants-in-Aid program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hein, J.W., Wolfe, G.V. & Blee, K.A. Comparison of Rhizosphere Bacterial Communities in Arabidopsis thaliana Mutants for Systemic Acquired Resistance. Microb Ecol 55, 333–343 (2008). https://doi.org/10.1007/s00248-007-9279-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-007-9279-1