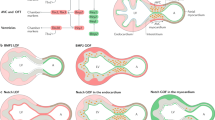
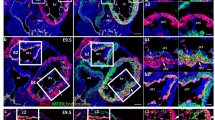
Abstract
The Notch pathway is an ancient, highly conserved signaling mechanism that participates in essential cell–cell communication events between adjacent cells. Mutations in Notch-signaling elements cause cardiac abnormalities in mice and humans, demonstrating an essential role for Notch in heart development. Studies with targeted mutant mice indicate that Notch signaling promotes the epithelial-to-mesenchyme transition that gives rise to the cardiac valve primordium, which later is sculpted into mature valves. During ventricular chamber development, the myocardium differentiates into two layers: an outer compact zone and an inner trabecular zone. Trabeculae provide a pumping function during early phases of ventricular development and contribute to the cardiac conduction system in the mature heart. Notch regulates the endocardium-to-myocardium signals that balance proliferation and differentiation of trabecular myocytes. Recent evidence demonstrates that defective NOTCH signaling leads to aortic valve degeneration in humans. Future research will be informative about the involvement of altered NOTCH signaling in chamber abnormalities and other cardiac disorders.



Similar content being viewed by others
References
Artavanis-Tsakonas S, Rand MD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284:770–776
Beis D, Bartman T, Jin SW et al (2005) Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 132:4193–4204
Ben-Shachar G, Arcilla RA, Lucas RV et al (1985) Ventricular trabeculations in the chick embryo heart and their contribution to ventricular and muscular septal development. Circ Res 57:759–766
Bettenhausen B, Hrabe de Angelis M, Simon D et al (1995) Transient and restricted expression during mouse embryogenesis of Dll1, a murine gene closely related to Drosophila delta. Development 121:2407–2418
Blank V, Kourilsky P, Israel A (1992) NF-kappa B and related proteins: Rel/dorsal homologies meet ankyrin-like repeats. Trends Biochem Sci 17:135–140
Bolos V, Grego-Bessa J, de la Pompa JL (2007) Notch signaling in development and cancer. Endocr Rev 28:339–363
Bruckner K, Perez L, Clausen H et al (2000) Glycosyltransferase activity of Fringe modulates Notch–Delta interactions. Nature 406:411–415
Chen F, Kook H, Milewski R et al (2002) Hop is an unusual homeobox gene that modulates cardiac development. Cell 110:713–723
Chen H, Shi S, Acosta L et al (2004) BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131:2219–2231
De Strooper B, Annaert W, Cupers P et al (1999) A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518–522
Del Amo FF, Smith DE, Swiatek PJ et al (1992) Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development 115:737–744
Del Monte G, Grego-Bessa J, Gonzalez-Rajal A et al (2007) Monitoring Notch1 activity in development: evidence for a feedback regulatory loop. Dev Dyn 236:2594–2614
Dunwoodie SL, Henrique D, Harrison SM et al (1997) Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development 124:3065–3076
Eldadah ZA, Hamosh A, Biery NJ et al (2001) Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet 10:163–169
Fehon RG, Kooh PJ, Rebay I et al (1990) Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell 61:523–534
Fischer A, Schumacher N, Maier M et al (2004) The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev 18:901–911
Fischer A, Steidl C, Wagner TU et al (2007) Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res 100:856–863
Garg V, Muth AN, Ransom JF et al (2005) Mutations in Notch1 cause aortic valve disease. Nature 437:270–274
Grego-Bessa J, Luna-Zurita L, del Monte G et al (2007) Notch signaling is essential for ventricular chamber development. Dev Cell 12:415–429
Hertig CM, Kubalak SW, Wang Y et al (1999) Synergistic roles of neuregulin-1 and insulin-like growth factor-1 in activation of the phosphatidylinositol 3-kinase pathway and cardiac chamber morphogenesis. J Biol Chem 274:37362–37369
Jenni R, Oechslin E, Schneider J et al (2001) Echocardiographic and pathoanatomical characteristics of isolated left ventricular noncompaction: a step towards classification as a distinct cardiomyopathy. Heart 86:666–671
Kokubo H, Miyagawa-Tomita S, Johnson RL (2005) Hesr, a mediator of the notch signaling functions in heart and vessel development. Trends Cardiovasc Med 15:190–194
Kokubo H, Miyagawa-Tomita S, Nakazawa M et al (2005) Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev Biol 278:301–309
Kokubo H, Miyagawa-Tomita S, Tomimatsu H et al (2004) Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ Res 95:540–547
Kokubo H, Tomita-Miyagawa S, Hamada Y et al (2007) Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development 134:747–755
Krebs LT, Xue Y, Norton CR et al (2000) Notch signaling is essential for vascular morphogenesis in mice. Genes Dev 14:1343–1352
Kurooka H, Honjo T (2000) Functional interaction between the mouse Notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem 275:17211–17220
Lardelli M, Dahlstrand J, Lendahl U (1994) The novel Notch homologue mouse Notch3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev 46:123–136
Lewis J (1998) Notch signaling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol 9:583–589
Lindsell CE, Shawber CJ, Boulter J et al (1995) Jagged: a mammalian ligand that activates Notch1. Cell 80:909–917
Logeat F, Bessia C, Brou C et al (1998) The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A 95:8108–8112
Loomes KM, Underkoffler LA, Morabito J et al (1999) The expression of Jagged1 in the developing mammalian heart correlates with cardiovascular disease in Alagille syndrome. Hum Mol Genet 8:2443–2449
McCright B, Gao X, Shen L et al (2001) Defects in development of the kidney, heart, and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491–502
McCright B, Lozier J, Gridley T (2002) A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 129:1075–1082
McDaniell R, Warthen DM, Sanchez-Lara PA et al (2006) NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 79:169–173
McLaughlin KA, Rones MS, Mercola M (2000) Notch regulates cell fate in the developing pronephros. Dev Biol 227:567–580
Milan DJ, Giokas AC, Serluca FC et al (2006) Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 133:1125–1132
Moloney DJ, Panin VM, Johnston SH et al (2000) Fringe is a glycosyltransferase that modifies Notch. Nature 406:369–375
Morel V, Lecourtois M, Massiani O et al (2001) Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol 11:789–792
Mumm JS, Schroeter EH, Saxena MT et al (2000) A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell 5:197–206
Nakagawa O, Nakagawa M, Richardson JA et al (1999) HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol 216:72–84
Nemir M, Croquelois A, Pedrazzini T et al (2006) Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res 98:1471–1478
Oka C, Nakano T, Wakeham A et al (1995) Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development 121:3291–3301
Panin VM, Papayannopoulos V, Wilson R et al (1997) Fringe modulates Notch–ligand interactions. Nature 387:908–912
Radtke F, Raj K (2003) The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer 3:756–767
Rechsteiner M (1988) Regulation of enzyme levels by proteolysis: the role of pest regions. Adv Enzyme Regul 27:135–151
Rentschler S, Zander J, Meyers K et al (2002) Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc Natl Acad Sci U S A 99:10464–10469
Runyan RB, Markwald RR (1983) Invasion of mesenchyme into three-dimensional collagen gels: a regional and temporal analysis of interaction in embryonic heart tissue. Dev Biol 95:108–114
Rutenberg JB, Fischer A, Jia H et al (2006) Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 133:4381–4390
Sakata Y, Kamei CN, Nakagami H et al (2002) Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc Natl Acad Sci U S A 99:16197–16202
Shawber C, Boulter J, Lindsell CE et al (1996) Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol 180:370–376
Shin CH, Liu ZP, Passier R et al (2002) Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110:725–735
Stollberger C, Finsterer J, Blazek G (2002) Left ventricular hypertrabeculation/noncompaction and association with additional cardiac abnormalities and neuromuscular disorders. Am J Cardiol 90:899–902
Swiatek PJ, Lindsell CE, del Amo FF et al (1994) Notch1 is essential for postimplantation development in mice. Genes Dev 8:707–719
Tamura K, Taniguchi Y, Minoguchi S et al (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr Biol 5:1416–1423
Timmerman LA, Grego-Bessa J, Raya A et al (2004) Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 18:99–115
Uyttendaele H, Marazzi G, Wu G et al (1996) Notch4/int-3, a mammary protooncogene, is an endothelial cell-specific mammalian Notch gene. Development 122:2251–2259
Venkatesh DA, Park KS, Harrington A et al (2008) Cardiovascular and hematopoietic defects associated with Notch1 activation in embryonic Tie2-expressing populations. Circ Res 103:423–431
Vuillemin M, Pexieder T (1989) Normal stages of cardiac organogenesis in the mouse: II. Development of the internal relief of the heart. Am J Anat 184:114–128
Watanabe Y, Kokubo H, Miyagawa-Tomita S et al (2006) Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development 133:1625–1634
Weinmaster G, Roberts VJ, Lemke G (1992) Notch2: a second mammalian Notch gene. Development 116:931–941
Wilson A, Radtke F (2006) Multiple functions of Notch signaling in self-renewing organs and cancer. FEBS Lett 580:2860–2868
Wu L, Aster JC, Blacklow SC et al (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet 26:484–489
Zhao YY, Sawyer DR, Baliga RR et al (1998) Neuregulins promote survival and growth of cardiac myocytes: persistence of ErbB2 and ErbB4 expression in neonatal and adult ventricular myocytes. J Biol Chem 273:10261–10269
Acknowledgments
The study was supported by grants SAF2007-62445 from the Spanish Ministry of Science and Innovation, S-BIO-0194/06 from the Regional Government of Madrid, RETICS 06/0014/0038 from the Spanish Ministry of Science and Innovation, and LSHM-CT-2005-018630 from the European Union 6th FP and the Fundación Rodríguez Pascual.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de la Pompa, J.L. Notch Signaling in Cardiac Development and Disease. Pediatr Cardiol 30, 643–650 (2009). https://doi.org/10.1007/s00246-008-9368-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00246-008-9368-z