Abstract
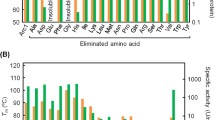
Understanding the origin and early evolution of proteins is important for unveiling how the RNA world developed into an RNA–protein world. Because the composition of organic molecules in the Earth’s primitive environment was plausibly not as diverse as today, the number of different amino acids used in early protein synthesis is likely to be substantially less than the current 20 proteinogenic residues. In this study, we have explored the thermal stability and RNA binding of ancestral variants of the ribosomal protein uS8 constructed from a reduced-alphabet of amino acids. First, we built a phylogenetic tree based on the amino acid sequences of uS8 from multiple extant organisms and used the tree to infer two plausible amino acid sequences corresponding to the last bacterial common ancestor of uS8. Both ancestral proteins were thermally stable and bound to an RNA fragment. By eliminating individual amino acid letters and monitoring thermal stability and RNA binding in the resulting proteins, we reduced the size of the amino acid set constituting one of the ancestral proteins, eventually finding that convergent sequences consisting of 15- or 14-amino acid alphabets still folded into stable structures that bound to the RNA fragment. Furthermore, a simplified variant reconstructed from a 13-amino-acid alphabet retained affinity for the RNA fragment, although it lost conformational stability. Collectively, RNA-binding activity may be achieved with a subset of the current 20 amino acids, raising the possibility of a simpler composition of RNA-binding proteins in the earliest stage of protein evolution.






Similar content being viewed by others
References
Akanuma S et al (2013) Experimental evidence for the thermophilicity of ancestral life. Proc Natl Acad Sci USA 110:11067–11072. https://doi.org/10.1073/pnas.1308215110
Akanuma S, Kigawa T, Yokoyama S (2002) Combinatorial mutagenesis to restrict amino acid usage in an enzyme to a reduced set. Proc Natl Acad Sci USA 99:13549–13553. https://doi.org/10.1073/pnas.222243999
Allmang C, Mougel M, Westhof E, Ehresmann B, Ehresmann C (1994) Role of conserved nucleotides in building the 16S rRNA binding site of E. coli ribosomal protein S8. Nucleic Acids Res 22:3708–3714. https://doi.org/10.1093/nar/22.18.3708
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Angyan AF, Ortutay C, Gaspari Z (2014) Are proposed early genetic codes capable of encoding viable proteins? J Mol Evol 78:263–274. https://doi.org/10.1007/s00239-014-9622-3
Ban N et al (2014) A new system for naming ribosomal proteins. Curr Opin Struct Bio 24:165–169. https://doi.org/10.1016/j.sbi.2014.01.002
Baumann U, Oro J (1993) Three stages in the evolution of the genetic code. Biosystems 29:133–141
Boussau B, Blanquart S, Necsulea A, Lartillot N, Gouy M (2008) Parallel adaptations to high temperatures in the Archaean eon. Nature 456:942–945. https://doi.org/10.1038/nature07393
Bridgham JT, Carroll SM, Thornton JW (2006) Evolution of hormone-receptor complexity by molecular exploitation. Science 312:97–101. https://doi.org/10.1126/science.1123348
Bridgham JT, Ortlund EA, Thornton JW (2009) An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature 461:515–519. https://doi.org/10.1038/nature08249
Busch F, Rajendran C, Heyn K, Schlee S, Merkl R, Sterner R (2016) Ancestral tryptophan synthase reveals functional sophistication of primordial enzyme complexes. Cell Chem Biol 23:709–715. https://doi.org/10.1016/j.chembiol.2016.05.009
Butzin NC, Lapierre P, Green AG, Swithers KS, Gogarten JP, Noll KM (2013) Reconstructed ancestral Myo-inositol-3-phosphate synthases indicate that ancestors of the thermococcales and thermotoga species were more thermophilic than their descendants. PLoS ONE 8:e84300. https://doi.org/10.1371/journal.pone.0084300
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Chyba CF (1990) Impact delivery and erosion of planetary oceans in the early inner solar system. Nature 343:129–133. https://doi.org/10.1038/343129a0
Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006) Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283–1287. https://doi.org/10.1126/science.1123061
Cleaves HJ 2nd (2010) The origin of the biologically coded amino acids. J Theor Biol 263:490–498. https://doi.org/10.1016/j.jtbi.2009.12.014
Collatz E, Wool IG, Lin A, Stöffler G (1976) The isolation of eukaryotic ribosomal proteins. the purification and characterization of the 40 S ribosomal subunit proteins S2, S3, S4, S5, S6, S7, S8, S9, S13, S23/S24, S27, and S28. J Biol Chem 251:4666–4672
Cornell CE et al (2019) Prebiotic amino acids bind to and stabilize prebiotic fatty acid membranes. Proc Natl Acad Sci USA 116:17239–17244. https://doi.org/10.1073/pnas.1900275116
Cornish-Bowden A, Cardenas ML (2017) Life before LUCA. J Theor Biol 434:68–74. https://doi.org/10.1016/j.jtbi.2017.05.023
Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM (2008) The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA 105:20356–20361. https://doi.org/10.1073/pnas.0810647105
Crick FH (1968) The origin of the genetic code. J Mol Biol 38:367–379
Cronin JR (1989) Origin of organic compounds in carbonaceous chondrites. Adv Space Res 9:59–64. https://doi.org/10.1016/0273-1177(89)90364-5
Cronin JR, Pizzarello S (1983) Amino acids in meteorites. Adv Space Res 3:5–18
Darwin C (1859) On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. John Murray, London
Davlieva M, Donarski J, Wang J, Shamoo Y, Nikonowicz EP (2014) Structure analysis of free and bound states of an RNA aptamer against ribosomal protein S8 from Bacillus anthracis. Nucleic Acids Res 42:10795–10808. https://doi.org/10.1093/nar/gku743
Doolittle WF (1999) Phylogenetic classification and the universal tree. Science 284:2124–2129
Edwards RJ, Shields DC (2004) GASP: gapped ancestral sequence prediction for proteins. BMC Bioinformatics 5:123. https://doi.org/10.1186/1471-2105-5-123
Eigen M, Schuster P (1977) The hypercycle. a principle of natural self-organization. part a: emergence of the hypercycle. Naturwissenschaften 64:541–565
Ferus M et al (2017) Formation of nucleobases in a miller-urey reducing atmosphere. Proc Natl Acad Sci USA 114:4306–4311. https://doi.org/10.1073/pnas.1700010114
Fournier GP, Gogarten JP (2010) Rooting the ribosomal tree of life. Mol Biol Evol 27:1792–1801. https://doi.org/10.1093/molbev/msq057
Francis BR (2013) Evolution of the genetic code by incorporation of amino acids that improved or changed protein function. J Mol Evol 77:134–158. https://doi.org/10.1007/s00239-013-9567-y
Furukawa R, Nakagawa M, Kuroyanagi T, Yokobori SI, Yamagishi A (2017) Quest for ancestors of eukaryal cells based on phylogenetic analyses of aminoacyl-tRNA synthetases. J Mol Evol 84:51–66. https://doi.org/10.1007/s00239-016-9768-2
Galtier N, Tourasse N, Gouy M (1999) A nonhyperthermophilic common ancestor to extant life forms. Science 283:220–221
Garcia AK, Kaçar B (2019) How to resurrect ancestral proteins as proxies for ancient biogeochemistry. Free Radic Biol Med 140:260–269. https://doi.org/10.1016/j.freeradbiomed.2019.03.033
Garcia AK, Schopf JW, Yokobori SI, Akanuma S, Yamagishi A (2017) Reconstructed ancestral enzymes suggest long-term cooling of earth’s photic zone since the Archean. Proc Natl Acad Sci USA 114:4619–4624. https://doi.org/10.1073/pnas.1702729114
Gaucher EA, Thomson JM, Burgan MF, Benner SA (2003) Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature 425:285–288. https://doi.org/10.1038/nature01977
Gaucher EA, Govindarajan S, Ganesh OK (2008) Palaeotemperature trend for precambrian life inferred from resurrected proteins. Nature 451:704–707. https://doi.org/10.1038/nature06510
Gaucher EA, Kratzer JT, Randall RN (2010) Deep phylogeny–how a tree can help characterize early life on earth. Cold Spring Harb Perspect Biol 2:a002238. https://doi.org/10.1101/cshperspect.a002238
Giacobelli VG et al (2022) In vitro evolution reveals noncationic protein-RNA interaction mediated by metal ions. Mol Biol Evol 39:msac032. https://doi.org/10.1093/molbev/msac032
Gilbert W (1986) Origin of life: the RNA world. Nature 319:618–618. https://doi.org/10.1038/319618a0
Granold M, Hajieva P, Toşa MI, Irimie FD, Moosmann B (2018) Modern diversification of the amino acid repertoire driven by oxygen. Proc Natl Acad Sci USA 115:41–46. https://doi.org/10.1073/pnas.1717100115
Gromiha MM, Oobatake M, Sarai A (1999) Important amino acid properties for enhanced thermostability from mesophilic to thermophilic proteins. Biophys Chem 82:51–67
Groussin M, Boussau B, Charles S, Blanquart S, Gouy M (2013) The molecular signal for the adaptation to cold temperature during early life on earth. Biol Lett 9:20130608
Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35:849–857. https://doi.org/10.1016/0092-8674(83)90117-4
Gumulya Y et al (2018) Engineering highly functional thermostable proteins using ancestral sequence reconstruction. Nat Catal 1:878–888. https://doi.org/10.1038/s41929-018-0159-5
Hanson-Smith V, Kolaczkowski B, Thornton JW (2010) Robustness of ancestral sequence reconstruction to phylogenetic uncertainty. Mol Biol Evol 27:1988–1999. https://doi.org/10.1093/molbev/msq081
Harms MJ, Thornton JW (2013) Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nat Rev Genet 14:559–571. https://doi.org/10.1038/nrg3540
Harris JK, Kelley ST, Spiegelman GB, Pace NR (2003) The genetic core of the universal ancestor. Genome Res 13:407–412. https://doi.org/10.1101/gr.652803
Higgs PG (2009) A four-column theory for the origin of the genetic code: tracing the evolutionary pathways that gave rise to an optimized code. Biol Direct 4:16. https://doi.org/10.1186/1745-6150-4-16
Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR (1993) Gene splicing by overlap extension. Methods Enzymol 217:270–279
Hug LA et al (2016) A new view of the tree of life. Nat Microbiol 1:16048. https://doi.org/10.1038/nmicrobiol.2016.48
Ikehara K, Omori Y, Arai R, Hirose A (2002) A novel theory on the origin of the genetic code: a GNC-SNS hypothesis. J Mol Evol 54:530–538. https://doi.org/10.1007/s00239-001-0053-6
Ilardo MA, Freeland SJ (2014) Testing for adaptive signatures of amino acid alphabet evolution using chemistry space. J Syst Chem 5:1. https://doi.org/10.1186/1759-2208-5-1
James JE, Willis SM, Nelson PG, Weibel C, Kosinski LJ, Masel J (2021) Universal and taxon-specific trends in protein sequences as a function of age. Elife. https://doi.org/10.7554/eLife.57347
Johnson DB, Wang L (2010) Imprints of the genetic code in the ribosome. Proc Natl Acad Sci USA 107:8298–8303. https://doi.org/10.1073/pnas.1000704107
Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S (2005) A universal trend of amino acid gain and loss in protein evolution. Nature 433:633–638. https://doi.org/10.1038/nature03306
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. https://doi.org/10.1038/nmeth.4285
Kandler O (1995) Cell wall biochemistry in archaea and its phylogenetic implications. J Biol Phys 20:165–169. https://doi.org/10.1007/bf00700433
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kędzior M, Garcia AK, Li M, Taton A, Adam ZR, Young JN, Kaçar B (2022) Resurrected Rubisco suggests uniform carbon isotope signatures over geologic time. Cell Rep 39:110726. https://doi.org/10.1016/j.celrep.2022.110726
Knauth LP, Lowe DR (1978) Oxygen isotope geochemistry of cherts from the onverwacht group (3.4 billion years), transvaal, south africa, with implications for secular variations in the isotopic composition of cherts. Earth Planet Sci Lett 41:209–222. https://doi.org/10.1016/0012-821X(78)90011-0
Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31:147–157. https://doi.org/10.1016/0092-8674(82)90414-7
Larralde R, Robertson MP, Miller SL (1995) Rates of decomposition of ribose and other sugars: implications for chemical evolution. Proc Natl Acad Sci USA 92:8158–8160
Lartillot N, Philippe H (2004) A bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21:1095–1109. https://doi.org/10.1093/molbev/msh112
Lartillot N, Brinkmann H, Philippe H (2007) Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol 7(Suppl 1):S4. https://doi.org/10.1186/1471-2148-7-s1-s4
Longo LM et al (2020) Primordial emergence of a nucleic acid-binding protein via phase separation and statistical ornithine-to-arginine conversion. Proc Natl Acad Sci USA 117:15731–15739. https://doi.org/10.1073/pnas.2001989117
Longo LM, Lee J, Blaber M (2013) Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein. Proc Natl Acad Sci USA 110:2135–2139. https://doi.org/10.1073/pnas.1219530110
Lupas AN, Alva V (2017) Ribosomal proteins as documents of the transition from unstructured (poly)peptides to folded proteins. J Struct Biol 198:74–81. https://doi.org/10.1016/j.jsb.2017.04.007
Mat WK, Xue H, Wong JT (2008) The genomics of LUCA. Front Biosci 13:5605–5613. https://doi.org/10.2741/3103
Mayer-Bacon C, Freeland SJ (2021) A broader context for understanding amino acid alphabet optimality. J Theor Biol 520:110661. https://doi.org/10.1016/j.jtbi.2021.110661
McDonald GD, Storrie-Lombardi MC (2010) Biochemical constraints in a protobiotic earth devoid of basic amino acids: the “BAA(-) world.” Astrobiology 10(10):989–1000. https://doi.org/10.1089/ast.2010.0484
Merkl R, Sterner R (2016) Ancestral protein reconstruction: techniques and applications. Biol Chem 397:1–21. https://doi.org/10.1515/hsz-2015-0158
Miller SL (1953) A production of amino acids under possible primitive earth conditions. Science 117:528–529
Miller SL, Lazcano A (1995) The origin of life–did it occur at high temperatures? J Mol Evol 41:689–692
Muller MM et al (2013) Directed evolution of a model primordial enzyme provides insights into the development of the genetic code. PLoS Genet 9:e1003187. https://doi.org/10.1371/journal.pgen.1003187
Nakamura E et al (2022) On the origin and evolution of the asteroid Ryugu: a comprehensive geochemical perspective. Proc Jpn Acad Ser B 98:227–282. https://doi.org/10.2183/pjab.98.015
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Nisbet EG, Sleep NH (2001) The habitat and nature of early life. Nature 409:1083–1091. https://doi.org/10.1038/35059210
Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289:920–930. https://doi.org/10.1126/science.289.5481.920
Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW (2007) Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317:1544–1548. https://doi.org/10.1126/science.1142819
Pace CN, Vajdos F, Fee L, Grimsley G, Gray T (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci 4:2411–2423. https://doi.org/10.1002/pro.5560041120
Patel BH, Percivalle C, Ritson DJ, Duffy CD, Sutherland JD (2015) Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nat Chem 7:301–307. https://doi.org/10.1038/nchem.2202
Pearce BKD, Pudritz RE, Semenov DA, Henning TK (2017) Origin of the RNA world: the fate of nucleobases in warm little ponds. Proc Natl Acad Sci USA 114:11327–11332. https://doi.org/10.1073/pnas.1710339114
Raymann K, Brochier-Armanet C, Gribaldo S (2015) The two-domain tree of life is linked to a new root for the Archaea. Proc Natl Acad Sci USA 112:6670–6675. https://doi.org/10.1073/pnas.1420858112
Rich A (1962) On the problems of evolution and biochemical information transfer. In: Kasha M, Pullman B (eds) Horizons in Biochemistry. Academic Press, NY, pp 103–126
Rinke C et al (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. https://doi.org/10.1038/nature12352
Rivera MC, Lake JA (1992) Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257:74–76
Robert F, Chaussidon M (2006) A palaeotemperature curve for the precambrian oceans based on silicon isotopes in cherts. Nature 443:969–972. https://doi.org/10.1038/nature05239
Rouet R et al (2017) Structural reconstruction of protein ancestry. Proc Natl Acad Sci USA 114:3897–3902. https://doi.org/10.1073/pnas.1613477114
Shibue R, Sasamoto T, Shimada M, Zhang B, Yamagishi A, Akanuma S (2018) Comprehensive reduction of amino acid set in a protein suggests the importance of prebiotic amino acids for stable proteins. Sci Rep 8:1227. https://doi.org/10.1038/s41598-018-19561-1
Shimojo M, Amikura K, Masuda K, Kanamori T, Ueda T, Shimizu Y (2020) In vitro reconstitution of functional small ribosomal subunit assembly for comprehensive analysis of ribosomal elements in E. coli. Commun Biol 3:142. https://doi.org/10.1038/s42003-020-0874-8
Solis AD (2019) Reduced alphabet of prebiotic amino acids optimally encodes the conformational space of diverse extant protein folds. BMC Evol Biol 19:158. https://doi.org/10.1186/s12862-019-1464-6
Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA (2005) Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet 37:630–635. https://doi.org/10.1038/ng1553
Thornton JW (2004) Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet 5:366–375. https://doi.org/10.1038/nrg1324
Trifonov EN (2000) Consensus temporal order of amino acids and evolution of the triplet code. Gene 261:139–151
Trudeau DL, Kaltenbach M, Tawfik DS (2016) On the potential origins of the high stability of reconstructed ancestral proteins. Mol Biol Evol 33:2633–2641. https://doi.org/10.1093/molbev/msw138
Vázquez-Salazar A, Lazcano A (2018) Early life: embracing the RNA world. Curr Biol 28:R220–R222. https://doi.org/10.1016/j.cub.2018.01.055
Weber AL, Miller SL (1981) Reasons for the occurrence of the twenty coded protein amino acids. J Mol Evol 17:273–284
Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF (2016) The physiology and habitat of the last universal common ancestor. Nat Microbiol 1:16116. https://doi.org/10.1038/nmicrobiol.2016.116
Wheeler LC, Lim SA, Marqusee S, Harms MJ (2016) The thermostability and specificity of ancient proteins. Curr Opin Struct Biol 38:37–43. https://doi.org/10.1016/j.sbi.2016.05.015
Wiener L, Schüler D, Brimacombe R (1988) Protein binding sites on E. coli 16S ribosomal RNA; RNA regions that are protected by proteins S7, S9 and S19, and by proteins S8, S15 and S17. Nucleic Acids Res 16:1233–1250. https://doi.org/10.1093/nar/16.4.1233
Williams PD, Pollock DD, Blackburne BP, Goldstein RA (2006) Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput Biol 2:e69. https://doi.org/10.1371/journal.pcbi.0020069
Williams TA, Foster PG, Cox CJ, Embley TM (2013) An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504:231–236. https://doi.org/10.1038/nature12779
Woese CR, Fox GE (1977) The concept of cellular evolution. J Mol Evol 10:1–6
Woese CR, Kandler O, Wheelis ML (1990) Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 87:4576–4579
Wolman Y, Haverland WJ, Miller SL (1972) Nonprotein amino acids from spark discharges and their comparison with the murchison meteorite amino acids. Proc Natl Acad Sci USA 69:809–811. https://doi.org/10.1073/pnas.69.4.809
Wong JT (1975) A co-evolution theory of the genetic code. Proc Natl Acad Sci USA 72:1909–1912
Yagi S et al (2021) Seven amino acid types suffice to create the core fold of RNA polymerase. J Am Chem Soc 143:15998–16006. https://doi.org/10.1021/jacs.1c05367
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. https://doi.org/10.1093/molbev/msm088
Yang Z, Rannala B (1997) Bayesian phylogenetic inference using DNA sequences: a Markov Chain Monte Carlo method. Mol Biol Evol 14:717–724
Yang Z, Kumar S, Nei M (1995) A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141:1641–1650
Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV (2008) The deep archaeal roots of eukaryotes. Mol Biol Evol 25:1619–1630. https://doi.org/10.1093/molbev/msn108
Akanuma S, Yamagishi A (2016) A strategy for designing thermostable enzymes by reconstructing ancestral sequences possessed by ancient life. Biotechnology of Extremophiles: Advances and Challenges, (ed Rampelotto PH), Springer, cham
Acknowledgements
The authors are grateful to Dr. Ryutaro Furukawa for assisting the phylogenetic analysis and ancestral sequence inference. This work was supported by JSPS KAKENHI (Grant Number 21H01200) and the Astrobiology Center Program of the National Institutes of Natural Sciences (Grant Number AB031007).
Funding
Japan Society for the Promotion of Science,21H01200,Satoshi Akanuma,National Institutes of Natural Sciences,AB031007,Satoshi Akanuma
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Belinda Chang .
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, F., Akanuma, S. Ancestral Sequence Reconstruction of the Ribosomal Protein uS8 and Reduction of Amino Acid Usage to a Smaller Alphabet. J Mol Evol 91, 10–23 (2023). https://doi.org/10.1007/s00239-022-10078-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-022-10078-w