Abstract
ATP synthase is a complex universal enzyme responsible for ATP synthesis across all kingdoms of life. The F-type ATP synthase has been suggested to have evolved from two functionally independent, catalytic (F1) and membrane bound (Fo), ancestral modules. While the modular evolution of the synthase is supported by studies indicating independent assembly of the two subunits, the presence of intermediate assembly products suggests a more complex evolutionary process. We analyzed the phylogenetic profiles of the human mitochondrial proteins and bacterial transcription units to gain additional insight into the evolution of the F-type ATP synthase complex. In this study, we report the presence of intermediary modules based on the phylogenetic profiles of the human mitochondrial proteins. The two main intermediary modules comprise the α3β3 hexamer in the F1 and the c-subunit ring in the Fo. A comprehensive analysis of bacterial transcription units of F1Fo ATP synthase revealed that while a long and constant order of F1Fo ATP synthase genes exists in a majority of bacterial genomes, highly conserved combinations of separate transcription units are present among certain bacterial classes and phyla. Based on our findings, we propose a model that includes the involvement of multiple modules in the evolution of F1Fo ATP synthase. The central and peripheral stalk subunits provide a link for the integration of the F1/Fo modules.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oxidative phosphorylation is the main source of ATP production in aerobic organisms. The redox reactions of the last step of aerobic respiration are performed by a series of evolutionary conserved protein complexes, or the electron transport chain (ETC). The main function of ETC is to produce a proton gradient across the cellular or mitochondrial membranes. ATP synthase utilizes this gradient to generate ATP (Mitchell 1961). Three types of ATP synthase have been identified: the A-, V-, and F-type ATPases. The A-type (A1Ao) ATPase exists in the archaea and a small number of bacteria (Ballmoos et al. 2008; Lewalter and Muller 2006). The V-type (V1Vo) ATPase exists in the eukaryotic cytoplasmic membranes (vacuoles) (Beyenbach and Wieczorek 2006). The F-type (F1Fo) ATP synthase is found in the mitochondria and chloroplasts of most eukaryotes and in the bacterial cells. The F1Fo ATP synthase has a range of structural features that distinguish them from the A- and V-type ATPases (Muller and Gruber 2003). A novel subfamily of F-type ATPases, named as “N-ATPase,” were identified in bacteria and some archaeal species (Methanosarcina acetivorans and M. barkeri). The N-type ATPase is thought to be acquired by a lateral gene transfer (Sumi et al. 1997; Dibrova et al. 2010). A distinctive property of the N-ATPase is believed to be its sodium translocating function, based on the presence of a full set of sodium-binding residues on their membrane-bound c subunits (Dibrova et al. 2010). The three ATPase types share a common ancestor, which has undergone structural and functional changes. The major subunits of A-type ATPases share about 50 and 25% sequence identity with V-type and F-type ATPases, respectively (Hilario and Gogarten 1998). Structurally, the A-, V-, and F-ATPases have three components: (i) a membrane-bound subunit, Ao/Vo/Fo, that includes the proton translocator, (ii) a connecting central stalk subunit, (iii) the catalytic subunit, A1/V1/F1 and (iv) the peripheral stalks, which are the least conserved component among the ATPases subtypes (Wang and Oster 1998; Gruber et al. 2001; Lee et al. 2010).
The bacterial F-type ATPase comprises 8 subunits (α3, β3, γ, δ, ε, a, b2, and c9−14) while the more complex mitochondrial F-ATPase consists of up to 17 subunits (α3, β3, γ, δ, ε, a, b2, c8−14, d, e, f, g, A6L, F6, I, K, and OSCP) (Buchanan and Walker 1996; Stock et al. 2000). The F1 complex is composed of an α and β hexameric subunit which includes the ATP catalytic site (Stock et al. 2000). The membrane-bound Fo component comprises a homomeric c-subunit ring and the ‘a’ subunit containing the proton translocator plus the b subunit (Lau et al. 2008; Rubinstein et al. 2003). The F1 and Fo components are connected by a central and a peripheral stalk. The mitochondrial F1Fo central stalk is formed by an elongated γ subunit and two regulator factors δ and ε subunits. Though universal, major differences exist between the exact subunit construction of F1Fo ATP synthase of prokaryotic vs. eukaryotic species. The bacterial peripheral stalk, for instance, is composed of δ and b2 subunits while the bovine version of the peripheral stalk is formed by Oligomycin sensitivity conferral protein (OSCP), F6, b, and d subunits, essential for the ATP synthase activity (Collinson et al. 1996). The N-terminal part of OSCP in mammals can interact directly with α subunit, and acts as a stator to prevent F1 hexamer rotation with the central rotary element (Rubinstein et al. 2003; Carbajo et al. 2005). The transfer of protons through Fo subunits causes the rotation of membrane-bound c-subunits and of the connected central stalk. ATP synthesis is achieved through a conformational change in the F1 as the γ subunit of the stalk rotates (Noji et al. 1997; Gibbons et al. 2000). The F1 hexamer can also hydrolyze ATP, which results in reversal of the direction of rotation of γ and the c-subunit ring. This process drives the protons against their concentration gradient, from the matrix to the intermembrane space of the mitochondria (Boyer 1997; Xing et al. 2005).
The two structurally and functionally distinct parts of the ATP synthase, F1 and Fo, have been suggested to comprise evolutionary modules (Walker 1998; Mulkidjanian et al. 2007). This view was supported by the evidence that the genes encoding F1 and Fo subunits are found gathered into separate transcription units in bacteria like Rhodospirillum rubrum (Falk and Walker 1988). Mulkidjanian et al. suggested an interesting evolutionary model that F1Fo ATP synthase originated from membrane protein translocases, which themselves evolved from RNA translocases. Following this model, the central stalk was originally the translocated protein trapped in the channel of the membrane translocase, and the peripheral stalk was formed to provide an elastic connection between the ancestral hexamer and the c-subunit ring (Mulkidjanian et al. 2007). A more recent study suggested that the assembly of yeast mitochondrial F1Fo ATP synthase is not a single linear process (as previously thought) but involves two separate regulated pathways (Rak et al. 2011).
In the present study, we generated the phylogenetic profiles of human mitochondrially targeted proteins by integrating the whole genome information of 759 different prokaryotic and eukaryotic species. We show for the first time that the F1-Fo ATP synthase subunits were consistently gathered into three major groups: F1, c-rings, and the remaining subunits of the Fo component. By analysis of transcription units from more than 2000 bacterial species, we also concluded (i) a constant order and conserved combination patterns of F1Fo ATP synthase subunits in bacterial genomes, (ii) F1 (or F1 plus b subunit) and Fo components were significantly gathered into different transcription units, and (iii) α/β and c-ring, respectively, in F1 and Fo components could be independent modules. Based on these results, we postulated that the F1Fo ATP synthase evolved from at least four modules, the α/β and c-ring and the remainder of the F1 and separately the Fo components. The central and peripheral stalk subunits may provide the evolutionary linkage for the combination of these different modules.
Materials and Methods
Phylogenetic Profiling of Human Mitochondria Proteins
Genomic sequences and annotation information were retrieved from the KEGG database (retrieved at 02/15/2014) (Kanehisa et al. 2006), and BLASTP (Camacho et al. 2009) was used for comparisons of protein sequences between human and the rest of 758 species. We constructed the phylogenetic profiles of each human protein sequence according the BLASTP E-value, and the “presence” and “absence” were recorded by using the E-value threshold 0.001.
We manually checked the homologs of F1Fo ATP synthase, 93.7% (4191/4473) were homologous subunits of F-type ATP synthase. For the total 213 homologous proteins (13 in Eukaryota, 36 in Bacteria, and 164 in Archaea) of V/A-type ATP synthase, they were either A subunit or B subunit which were homologs of α and β subunits, respectively. Bedsides, these detected A/B subunits were from species with complete or partial lack of F-type ATP synthase, demonstrating that we have detected accurate homologs of human F1Fo ATP synthase without messing up with V/A-type ATPase paralogs. We proposed six independent methods to measure the distance between pairs of genes (Glazko and Mushegian 2004; Kensche et al. 2008), namely, Euclidean, Manhattan, Jaccard, tau, mutual information, and Pearson absolute anti-correlation distance. A total of 1006 human mitochondrial proteins were chosen from MitoCarta database (Pagliarini et al. 2008) for hierarchical cluster analysis, and grouped number was set as 5, 10, 15, 20, 25, 30, 35, 40, 45, and 50. Moreover, PCA was applied for the group analysis of mitochondrial F1Fo ATP synthase proteins.
Hierarchical Cluster Methods Used in Phylogenetic Profile Analysis
To get robust cluster analysis in phylogenetic profiling analysis, we applied a variety of hierarchical cluster methods (Glazko and Mushegian 2004; Kensche et al. 2008). Each pair of human mitochondrial proteins was denoted as two vectors containing either 0 (“absence”) and 1 (“presence”), for example \(X,Y\in \{\text{0,1}{\}}^{n}\).
The \({L}_{p}\)-norms is
where \(n\) is the total number of species used in phylogenetic profiling; \(p=1\) is the Manhattan distance; and \(p=2\) is the Euclidean distance.
Jaccard distance is defined from co-occurrence of presences
Mutual information is used to measure distance (Korber et al. 1993; Huynen et al. 2000),
where \(p \left(x\right)\) is the probability that a symbol (here 0 or 1) appears in the gene vector \(X\)regardless of the probability of a symbol combination appearing in gene vectors \(X\) and Y. The mutual information distance is defined as \({d}_{M}=1-I (X;Y)\).
The Kendall tau rank distance is defined as
where \(\tau 1\) and \(\tau 2\) are the rankings of elements in \(X\) and \(Y\); \(P\) is the paired combination in \(\tau 1\) and \(\tau 2\). If \(i\) and \(j\) are in the same order, \({\overline{K}}_{i,j}=0\); otherwise, \({\overline{K}}_{i,j}=1\).
We used the Pearson correlation coefficient
where \(\overline{X}\) and \(\overline{Y}\) are the sample means of \(X\) and Y, and \({S}_{X}\) and \({S}_{Y}\) are the sample standard deviations of \(X\) and \(Y\). The Pearson absolute anti-correlation distance is \({d}_{P}=1-\left|\text{c}\text{o}\text{r} \left(X,Y\right)\right|\).
We applied the Euclidean distance measurement to generate the distance tree (Fig. 2a–c) with phylogenetic profiles of human mitochondria proteins, in which BLASTP E-value 0.001 was used as the threshold. We then used the threshold 0.1, 0.01, and 0.0001 to validate the distance tree (Suppl. Fig. 2). The group number of PCA was set as 2, 3 (Suppl. Fig. 3), and 4 (Fig. 2d) to cluster the mitochondria F1Fo ATP synthase subunits.
Identification of Evolutionarily Conserved Modules
We applied the clustering by inferred models of evolution (CLIME) algorithm with default phylogenetic tree (Bick et al. 2012) and parameters to identify evolutionarily conserved modules (ECMs) of F1Fo ATP synthase subunits (Li et al. 2014). The input phylogenetic profile using human as the reference was prepared as described in CLIME, and the same threshold (BLASTP E-value < 0.001) was set. The single “Prokaryotes” was generated from 696 prokaryotic species, in which the “presence” was assigned if more 5% homologs were detected.
Prediction of Bacterial Transcription Units
Orthology of bacterial F1Fo ATP synthase genes was provided in KEGG Orthology database (http://www.genome.jp/kegg/ko.html). We included 2248 different bacterial strains, and retrieved their transcription unit information from the BioCyc database (http://biocyc.org/).
Bacterial transcription unit data of F1Fo ATP synthase genes were retrieved from BioCyc (retrieved on 04/10/2014), and most of them were predicted by computational methods (Romero and Karp 2004). We improved the prediction method by combining the continuous transcription units if they were in the same transcription direction and had no interrupted gene between them (Fig. 3b and Suppl. File 1). We at the same time reserved the raw transcription unit data (Suppl. File 2) and used the parallel analysis procedure (Suppl. Fig. 4).
Bacterial Transcription Unit Analysis
The frequency of transcription unit modules presented in Fig. 3b was generated by mapping the transcription unit data to the most common gene order “ε-β-γ-α-δ-b-c-a” without considering the transcription direction. We defined the standard cTU as “ε-β-γ-α-δ-b-c-a” because this long ordered transcription unit was observed in most species in which all the F1Fo ATP synthase genes were maintained together in the same transcription units. In this process, we merged the homology of the synthase genes if they were continuously arranged in the same transcription unit; for example, we transferred the transcription unit “ε-β-γ-α-δ-b-b′” to “ε-β-γ-α-δ-b,” as the gene order was the most important feature with which we were concerned.
We used the upper confidence intervals of the binomial distribution as the cutoff line (Fig. 3b and Suppl. Fig. 4)
where \(p\) is the possibility to choose one pair of cut points as denoted in Fig. 3a, here \(p=1/C\left(9, 2\right)=1/36\), \({Z}_{\alpha }\) is the normal distribution quantile with \(\alpha =0.05\), and \(N\) is the total number of transcription units.
The δ/b Rosetta Stone was defined by appearance of the b and δ subunit motifs in the same protein sequence. The KEGG SSDB (Sequence Similarity DataBase, http://www.kegg.jp/kegg/ssdb/) was used to validate the δ/b Rosetta Stone (Suppl. Fig. 5).
Phylogenetic Tree
The 16S rDNA sequences were retrieved and used to reconstruct phylogenetic tree of bacteria species in transcription unit analysis. The bacterial class/phylum tree was constructed from concatenating 16S rDNA sequences in each class or phylum.
Alignments were generated using the MAFFT program (Katoh and Standley 2013), and the phylogenetic tree was constructed by setting the maximum likelihood in RAxML (Randomized Axelerated Maximum Likelihood) program (Stamatakis 2014) with default parameters. The transcription units were combined with the phylogenetic tree visualized in the iTOL (Interactive Tree Of Life) tool (Letunic and Bork 2011). The root of phylogenetic trees was determined at Thermotogae as was suggested by Battistuzzi and Hedges (2009). In the phylogenetic trees showing combination of transcription units and chimera fusions in bacterial phyla or classes, Fervidobacterium nodosum was set as the outer group.
Statistical Analysis
All the statistical analysis was conducted using the open-source R Project (http://www.r-project.org/).
Results
Construction of the Phylogenetic Profile of Mitochondrial Proteins
Based on the assumption that functionally related proteins are likely to evolve in a correlated manner, phylogenetic profiling has been used to predict functional and structural interactions between proteins (Pellegrini et al. 1999). To perform a comprehensive phylogenetic profile analysis across a wide range of eukaryotic and prokaryotic taxa, we considered all fully sequenced genomes and included at least one species from every class according to the taxonomy information provided in KEGG database (Kanehisa et al. 2006). The final list included 759 species including 82 Archaeal, 614 Bacterial, and 63 Eukaryotic species (Suppl. Table 1). The phylogenetic profiles were built for a total of 1006 human genes including the nuclear and mitochondrial protein-coding sequences for the mitochondrial proteins annotated by the MitoCarta database (Pagliarini et al. 2008) (Suppl. Table 2).
Subunits of Mitochondrial F1Fo ATP Synthase are Clustered in Distinct Groups
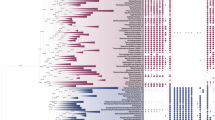
To accurately assess the evolutionary correlation of mitochondrial F1Fo ATP synthase subunits, we chose a two-step approach. First, we performed a hierarchical cluster analysis to examine whether the mitochondrial proteins would be grouped into different categories. By this analysis, the proteins that clustered into the same group need to be gained and lost together, based on the phylogenetic profiling data. Then, to get robust results, we applied 6 independent methods, Euclidean, Manhattan, Jaccard, Kendall tau, mutual information, and Pearson absolute anti-correlation distance, to calculate the distance within each gene pair. Using each method, we generated hierarchical groups of 5, to 50 (in increments of 5) mitochondrial protein-coding genes (Suppl. Table 3). Using the Euclidean distance and Manhattan distance methods, we revealed two distinct clusters of ATP synthase subunits. The catalytic subunits of the synthase, α (ATP5A1) and β (ATP5B), the central and lateral stalk subunits, γ (ATP5C1) and OSCP (ATP5O) (respectively), in addition to the a subunit of Fo clustered together (red). The rest of the subunits clustered into a second group (green). The use of Jaccard method placed the three homologs of the c subunit (ATP5G1, ATP5G2, and ATP5G3) in the first cluster (Fig. 1a).
Groups of mitochondrial F1Fo ATP synthase from mitochondrial proteins. Hierarchical clusters of phylogenetic profiles using 758 (a) or 490 randomly chosen (b) species with independent distance measurements: Euclidean, Manhattan, Jaccard, Kendall tau, mutual information, and Pearson absolute anti-correlation distance. Group number is set to 10, 25, or 40, and distinct groups are marked with different colors. (Color figure online)
To test the specificity of group members, we gradually increased the number of hierarchical groups from 5 to 50 (Suppl. Table 3). Interestingly, we detected two additional subgroups which had not been reported previously. The first subgroup consists of the α3β3 hexamer (purple), which contained the catalytic site of the F1 subunit. This appeared as an independent subgroup even when a small group number (10) was set with the methods of Kendall tau, mutual information, and Pearson absolute anti-correlation distance. Upon increasing the group number to 40, another subgroup containing the three homologs (ATP5G1/G2/G3) of the mitochondrial inner membrane-bound c-subunit appeared as a separately evolved group from the other Fo components according to Kendall tau and Pearson absolute anti-correlation distance methods (Fig. 1a, yellow).
To verify that this pattern did not appear as a result of using the specific species list chosen for the profiling analysis, we randomly selected 490 species including 50 Archaeal, 400 Bacterial, and 40 Eukaryotic species (Suppl. Table 4). The same analysis procedures, cluster methods, and hierarchical group numbers were applied to the newly generated phylogenetic profiles (Fig. 1b). These results strongly confirmed placement of the synthase subunits into four distinct clusters (Fig. 1b and Suppl. Table 5).
The Phylogenetic Profiles of the Subunits Within Each Evolutionary Module are Tightly Correlated
The finding that the α3β3 hexamer and the c-subunit homologs comprised distinct evolutionary modules prompted further investigation into the correlation of the genes within the evolutionary modules of the mitochondrial ATP synthase. We generated a distance tree and a correlation matrix from the evolutionary profile data of genes coding for 17 synthase subunits (Fig. 2a). The F1 subunits α and β (purple), γ and OSCP (red), appeared in the same cluster (Fig. 2b). The appearance of two different subgroups may be due to the fact that α/β or homologous A/B subunits in V/A-type ATPase subunits are present in all three domains of life, whereas γ and OSCP subunits exist in most eukaryotic and bacterial species but no significant homologs found in Archaea (Fig. 2a). The pairing of the subunits was confirmed by the high Pearson correlation coefficient value (0.54 for α/β and 0.70 for γ/OSCP) (Fig. 2c). In species harboring entire F1Fo ATP synthases, we detected the corresponding homologs of α and β subunits (Suppl. Fig. 1A). At the same threshold (BLASTP E-value smaller than 0.001), in organisms (mostly in Archaea and some Eukaryota and Bacteria) only possessing V/A-type ATPase or partial F-type ATPase, in the absence of α and/or β, the corresponding A/B subunits were chosen as the homologs (Suppl. Fig. 1B). The widely distributed homologs of α/β made them distinct from the rest of the subunits of the F1Fo ATP synthase. This subgroup was therefore considered as a separate evolutionary module.
Cluster analysis for human mitochondrial F1Fo ATP synthase subunits. a The phylogenetic profile for human 17 subunits of the F1Fo ATP synthase clustered both by species (columns) and genes (rows). The top color bar indicates the three domains of life, and the left color bar represents the corresponding F1 and Fo subunits arranged in a cluster dendrogram. The blue and gray bars in the phylogenetic profiles indicate presence or absence in a given species, respectively. b Distance tree of mitochondrial F1Fo ATP synthase subunits generated by Euclidean distance with the group number set to 6. Different colors are used to indicate identified clusters. c Correlation matrix of mitochondrial F1Fo ATP synthase subunits. The Pearson correlation coefficients of paired genes are marked in numbers and color-coded from blue to red. The left cluster dendrogram indicates the Euclidean distance between subunits of F1 and Fo marked with red and green, respectively. d Principal component analysis (PCA) plot of the 17 subunits of F1Fo ATP synthases with a cluster number of 4. Identified groups are shown with different colors and shapes. e The evolutionarily conserved modules (ECMs) of human F1Fo ATP synthase. Blue and gray blocks indicate present and absent homologs, respectively. (Color figure online)
Although all Fo subunits, c (ATP5G1/G2/G3), d (ATP5H), b (ATP5F1), g (ATP5L), f (ATP5J2), e (ATP5I), F6 (ATP5J), and A6L (ATP8) belonged to the same clade (Fig. 2b), the c-ring subunits (yellow) had a distinct pattern of phylogenetic profiles and high correlation coefficients (> 0.93 for each pair), which confirmed their existence as a separate subgroup (Fig. 2b, c). Changing the threshold of BLAST E-value (from 0.1 to 0.0001) for generating the distance trees did not alter the results and the α/β, γ/OSCP, and c-ring subgroups were consistently observed (Suppl. Fig. 2). The use of principal component analysis (PCA) highlighted the presence of four evolutionary modules: one comprised the c-ring subunits (yellow); the second included the α and β subunits (purple); the third and fourth included the known F1 (red) and Fo (green) modules each comprised the rest of the respective subunits of each compartment (Fig. 2d and Suppl. Fig. 3).
To further confirm these cluster modules, we applied clustering by inferred models of evolution (CLIME) method (Li et al. 2014) to the phylogenetic profiles. By combining the phylogenetic tree and profiles, CLIME can identify evolutionarily conserved modules (ECMs) from protein complexes and pathways. Three ECMs, α/β, OSCP/γ, and the c-rings, were detected (Fig. 2e), confirming the presence of multiple evolutionary modules for the F1Fo ATP synthase. We noticed that δ (ATP5D) and ε (ATP5E) subunits, components of central stalk, were clustered into separate groups (Fig. 2b), mainly due to their different gain and loss patterns. The absence of ε subunit in many protists (Fig. 2e) may have resulted from the incomplete genes annotation or different evolutionary scenarios for δ and ε subunit (Koumandou and Kossida 2014).
The Bacterial F1/Fo Subunits are Gathered into Different Modules
The bacterial F1Fo ATP synthase comprises 8 subunits. Subunits α, β, γ, δ, and ε belong to the F1 component, and the Fo component consists of the a, b (and b′), and c subunits (Deckers-Hebestreit and Altendorf 1996). While the genes coding for all eight subunits are arranged in the same operon and transcribed together in some bacteria, including Escherichia coli (Nielsen et al. 1984), the genes coding for the F1 and Fo components are clustered into separate operons in the genomes of other bacterial species including R. rubrum (Falk and Walker 1988). This raised the possibility that the F1Fo ATP synthase has evolved from two different modules (Falk and Walker 1988). To further investigate this premise, we performed a comprehensive analysis of transcription units containing the genes coding for bacterial F1Fo ATP synthase. For this analysis, we included a dataset of putative transcription units from 2248 strains or substrains of bacteria, covering 568 different genera, predicted by computational methods (Suppl. File 1) (Romero and Karp 2004).
As anticipated, a large percentage of species contained all eight subunits in the same transcription unit (cTU, complete transcription unit). Interestingly, the observation that ATP synthase genes were divided into different transcription units (iTU, incomplete transcription unit) was not rare. In addition to the iTUs reported in Rhodospirillum and Synechococcus, the genera Rickettsia and Methylobacterium contained “ε-β-γ-α-δ” and “b-c-a” iTUs, whereas the “ε-β-γ-α-δ-b,” “c,” and “a” iTUs were found in the genus Helicobacter (Suppl. File 1 and Suppl. Table 6). With a few exceptions, genes in most cTUs follow a constant order as “ε-β-γ-α-δ-b-c-a,” confirmed by experimental data from E. coli (Gay and Walker 1981), Bacillus subtilis (Santana et al. 1994), and Clostridium pasteurianum (Das and Ljungdahl 2003). The order of genes in iTUs, according to our data and previous studies (McCarn et al. 1988; Borghese et al. 1998), was similar to the order of genes in cTUs, suggesting that multiple distinct evolutionary modules, comprised a combination of ordered iTUs, exist within the bacterial F1Fo ATP synthase.
To test this hypothesis, we calculated the frequency of iTUs in various bacterial genomes. We used the most common gene order “ε-β-γ-α-δ-b-c-a” (in either direction) as the standard order, and then mapped each iTU and cTU accordingly (Fig. 3a). We neglected the transcriptional direction, which meant the “ε-β-γ-α-δ” and the “δ-α-γ-β-ε” were treated as the same iTU. As expected, we observed that standard order had the highest (34.6%) frequency, and the Fo module (“b-c-a”) and the F1 module (“ε-β-γ-α-δ”) had the second (9.8%) and the fifth (6.5%) highest frequencies, respectively. We identified three novel combinations, i.e., “ε-β” plus “γ-α” plus “δ-b-c-a,” “ε-β-γ-α-δ-b,” plus “c-a,” and “ε-β-γ-α-δ-b” plus “c” plus “a” (Fig 3b, c) in addition to the previously reported combination patters (“ε-β-γ-α-δ” plus “b-c-a” and “ε-β” plus “γ-α-δ-b-c-a”). The frequencies of nine modules were significantly higher than the upper confidence interval (0.03 with the threshold of 0.05) (Fig. 3b), suggesting that the modules are highly unlikely to be chosen randomly in bacterial genomes.
Transcription unit modules of bacterial F1Fo ATP synthase. a The standard transcription units with cut points. b The frequency of transcription unit modules mapped to the standard one in A. Red dots denote that transcription unit modules are significantly higher than the upper confidence interval. The green dots are the modules which did not reach statistical significance. The dashed gray line marks the upper confidence interval (0.03) with the threshold 0.05. c Representative patterns of transcription units detected in bacterial genomes. The F1Fo ATP synthase genes are marked with different colors and shapes. The schematic patterns do not indicate the length of the genome or the actual transcription direction. (Color figure online)
Prediction of the bacterial transcription units based on the BioCyc database (http://biocyc.org/) validated most of the identified modules, comprising the F1Fo ATP synthase subunits (Suppl. Fig. 4 and Suppl. File 2). Interestingly, a Rosetta Stone protein (a fusion protein generated by the combination of separate proteins) of δ and b subunits was observed in Gordonia genus and some Mycobacterium species (Fig. 3c and Suppl. Fig. 5). The identified δ/b Rosetta Stone was located in the middle of transcription units, between the α and another single b subunit (Suppl. Table 7). The δ/b Rosetta Stone is likely to result from a fusion event in the transcription unit, in which the δ and the two heterologous b subunit genes were arranged in a sequential order. Since iTUs such as “γ-α-δ-b-b′-c-a” and “ε-β-γ-α-δ-b-b′” were observed, it is likely that this fusional combination was a common way to generate multiple-domain proteins (Pasek et al. 2006; Wang and Caetano-Anolles 2009). The δ/b Rosetta Stone was unlikely to be directly generated by integrating the “b′-b-c-a” and “ε-β-γ-α-δ,” since these two kinds of iTUs were simultaneously found in some bacterial genomes.
Our transcription unit analysis revealed that the bacterial F1Fo ATP synthase genes were included in the same transcription unit in many bacterial genomes and they were arranged in a highly conserved order. The proximity of the F1Fo ATP synthase genes in transcription units is a dominant trait among various bacterial genomes, most likely to have been caused by positive selection. Further analysis of the diverse pool of bacterial species revealed at least five combinations of transcription units which contained parts of ATP synthase genes, arranged in a similar order to that of the standard cTU (Fig. 3c).
Transcription Units of F1Fo ATP Synthase Genes Were Conserved Among Bacterial Classes
The formation of transcription units containing multiple genes or operons has been ascribed to the process of horizontal gene transfer (HGT), in which genes close to each other are transferred together from one species to another. These genes may be functionally unrelated at first, but random genome rearrangements would move them closer and they would then transfer together. An operon then is formed after the deletion of intervening DNA sequences between adjacent genes (Lawrence and Roth 1996). An alternative model posits that operon formation is driven by co-regulation of genes instead of HGT. According to this model, individually transcribed genes with similar expression and regulation patterns integrate into one transcription unit to reduce the amount of regulatory information (Price et al. 2005).
To confirm this model, we constructed a phylogenetic tree based on the bacterial 16S rDNA sequences, and then mapped the transcription units modules to each branch (Fig. 4a). Interestingly, we did not observe a random distribution of different types of transcription and bacteria in the same class inclined to have the same transcription unit combination for the ATP synthase genes. For example, a considerable proportion of transcription units in the Gammaproteobacteria (94.0%), Betaproteobacteria (98.6%), Actinobacteria (78.6%), and many other bacterial classes (or phyla) were standard cTUs, and the genes in cTUs were generally arranged in a constant order. The phyla Bacteroidetes (87.8%) and Cyanobacteria (89.7%) had a combination of two distinct iTUs “ε-β” and “γ-α-δ-b-c-a,” while Deltaproteobacteria (76.3%) had “ε-β-γ-α-δ-b” and “c-a”; Alphaproteobacteria (89.1%) had “ε-β-γ-α-δ” and “b-c-a” and Epsilonproteobacteria (97.8%) had a triple transcription units “ε-β-γ-α-δ-b,” “c,” and “a” (Fig. 4b and Suppl. Table 8). The constant order of genes suggests that HGT may not be the major force in evolution of the F1Fo ATP synthase operons.
Dominant patterns of transcription units in bacterial classes and phyla. a A maximum-likelihood tree based on the bacterial 16S rDNA sequences. Bacterial classes and phyla are shown with distinct colors. A gradient of red color is used to represent the repeated transcription unit (modules) in the same genus. b A class/phylum phylogenetic tree combined with the dominant patterns of transcription units in each bacterial class and phylum. The histogram indicates the percentage of dominant patterns. (Color figure online)
To further investigate the possible evolutionary routes of bacterial F1Fo ATP synthase, we then mapped predicted transcription units into reconstructed phylogenetic trees in each bacterial phylum/class. Interestingly, the standard long transcription unit “ε-β-γ-α-δ-b-c-a” was not the only version of cTU. For example, the phylum Bacteroidetes possessed one minor cTU “β-ε-a-c-b-δ-α-γ” (Fig. 5a). In the class Bacilli, we found that the standard cTUs only occupy 65.9% of the 396 different species, while more than 30.8% of genomes contained another type of cTU, “ε-β-γ-α-δ-b-a-c” (Fig. 5b). Furthermore, phylogenetic analysis showed these minor cTUs concentrated in the species with rather close phylogenetic relationships, including the genera Bacteroides and Prevotella in Bacteroidetes (Fig. 5a) and genera Lactococcus and Streptococcus in Bacilli (Fig. 5b), indicating a relatively ancient gene inheritance. The minor cTUs could have been formed by the fusion of incomplete transcription modules, as the species adjacent to “β-ε-a-c-b-δ-α-γ” contained a combination pattern of “β-ε” plus “a-c-b-δ” plus “α-γ” or “β-ε” plus “a-c-b-δ-α-γ” (Fig. 5a). The minor cTU “ε-β-γ-α-δ-b-a-c” was possibly formed by the combination of “ε-β-γ-α-δ-b” and an inverse order of “c-a,” which took a large percentage in the class Deltaproteobacteria (Fig. 5b). Another novel trait in bacterial species was the presence of chimera fusions consisting of partial subunits in addition to the entire F1Fo ATP synthase. One of the frequent chimera fusions was “β-ε-a-c-b-α-γ” that was widely present in the classes Deltaproteobacteria (Fig. 5c), Betaproteobacteria (Fig. 5d), Alphaproteobacteria (Suppl. Fig. 6A), Gammaproteobacteria (Suppl. Fig. 6C), the phylum Cyanobacteria (Suppl. Table 6), and Chlorobi (Suppl. Table 6). The order of this kind of chimera fusion was consistent with the previously identified N-ATPase (Sumi et al. 1997; Dibrova et al. 2010), which is thought to be an early divergence from the ancestral ATPase (Dibrova et al. 2010). Other types of chimera fusion such as “ε-β-γ-α-δ-c” in Bacilli (Fig. 5b) and “ε-β-γ-α-b-c-a” in Clostridia (Suppl. Fig. 6B) were also detected. Considering these non-random modules (Fig. 3b) and other modules such as the F1 module (“ε-β-γ-α-δ”), “c,” “ε-β,” and “a-c-b,” the presence of chimera transcription units was not surprising and corroborating the modular evolution of F1Fo ATP synthase. Of these minor cTU and chimera fusions, only one special type of chimera fusion (“α-β”) in the phylum Tenericutes (Suppl. Fig. 6D, F) could not be predicted by direct combination of iTUs.
Distribution of minor cTUs and chimera fusions supporting the modular evolution. a–d The reconstructed phylogenetic tree combined with predicted transcription units containing F1Fo ATP synthase subunits in Bacteroidetes, Bacilli, Deltaproteobacteria, and Betaproteobacteria, respectively. Species in one genus are marked with the same color in bars and tree branches. (Color figure online)
Discussion
The F1Fo ATP synthase is a universal enzyme responsible for coupling ion movement across mitochondrial, chloroplast, or bacterial membranes with ATP synthesis. The structure of the synthase comprises a catalytic core (F1) and a membrane-bound subunit (Fo). The two functionally distinct subunits have been proposed to have evolved from functionally unrelated proteins (Mulkidjanian et al. 2007; Falk and Walker 1988; Rak et al. 2011). While the F1 subunit was proposed to have evolved from an ATP-dependent helicase, the Fo has been suggested to have evolved from a passive ion channel (Gomis-Ruth et al. 2001). The formation of the assembly intermediates (Rak et al. 2011; Havlickova et al. 2010) has suggested a more complex evolutionary process that may underlie the functional complexity of the synthase subunits.
The Phylogenetic Profiles of the F1Fo ATP Synthase Subunits Support Modular Evolution
Our analysis of the phylogenetic profiles of the synthase subunits revealed two previously unknown evolutionary modules, the c-ring and α3β3 hexamer, in addition to the previously proposed F1 an Fo units (Walker 1998). The presence of the additional two modules, deduced from the phylogenetic profile correlations, could simply be attributed to the high sequence similarity between subunits within the same module. For instance, the amino acid sequences of α and β subunits show a high sequence similarity (BLASTP E-value < 0.0001 and 26% sequence identities) in both human and E. coli. This pair of subunits is known to have duplicated before the last universal common ancestor and have been universally inherited as a pair since then (Iwabe et al. 1989; Shih and Matzke 2013). Alternatively, the presence of the modules could be explained through the functional correlation and formation of the assembly intermediates. This notion is supported by the studies showing that the F1 assembly is independent from the rest of the complex (Tzagoloff 1969) and that the oligomerization of the c subunits forms a bona fide assembly intermediate (Rak et al. 2011). Furthermore, it has been shown that the disruption of the F1Fo assembly results in accumulation of the c-subunit ring structure as an independent subcomplex (Havlickova et al. 2010). The finding that the c-subunit ring constitutes an independent evolutionary module, in combination with our recent results ascribing a role for the c-subunit in formation of a non-selective ion channel in the mitochondrial inner membrane (Alavian et al. 2014) suggest that the ancestral function of the c-subunit evolutionary module, as a passive ion channel, may be conserved throughout evolution.
Modular Evolution of F1Fo ATP Synthase Involved Fusion of Incomplete Transcription Units
Although a large number of bacterial classes possess a long and highly conserved transcription unit (cTU), our results strongly suggest that the bacterial F1Fo ATP synthase genes evolved from several distinct transcription unit modules. Gene order in transcription units is generally conserved among the phylogenetically close species, but this conservation is rapidly lost over a short period of evolutionary time (Tamames 2001). The order of the ATP synthase standard cTU, “ε-β-γ-α-δ-b-c-a,” however, remained rather stable even in distant bacterial classes (Fig. 4b), which argued against the involvement of horizontal gene transfer in this process. The constant order of ATP synthase genes in their genomic context is not a unique occurrence. The genes in ribosome, glutamate ABC transport system, and flagellum-related operons are also arranged in a highly ordered manner. The constant order is known to result from a conservative rearrangement of discrete sets of genes, which may be functionally related, have physical interaction, or share similar transcriptional regulatory elements (Tamames 2001; Lathe et al. 2000). Alternatively, these observed iTUs may have appeared due to the splits within the cTU, possibly by bacterial genome rearrangement. This scenario, however, seems unlikely since the frequency of genome rearrangement is high during evolution and the splitting of cTUs may interrupt the continuously arranged genes resulting in broken transcription units or replacement of native genes with foreign genes (Price et al. 2006). As the integrity of the transcription unit is of great importance for keeping the stoichiometric accuracy of the ATP synthase holoenzyme, any unexpected interruption would cause disadvantageous influences on the organisms. The highly ordered ATP synthase genes, therefore, seem to have been under a positive selection that forced them together. This was confirmed by the example of fusion of δ and b subunits of ATP synthase to a Rosetta stone.
The modular evolution of the synthase complex is further supported by the presence of the other types of cTUs and chimera fusion products in some bacteria. The presence of these unusual fusion products was predicted since it would be implausible to form long ordered transcription units by limited genome rearrangements during evolution. Further phylogenetic analysis of these minor cTUs supported the fusion model. In the phylum Bacteroidetes, bacteria species concentrated in genera Bacteroides and Prevotella had the minor cTU “β-ε-a-c-b-δ-α-γ” (Fig. 5a). At the same time, the combination patterns of “β-ε” plus “a-c-b-δ” plus “γ-α” were found at the nearby branches, which could be the resources formatting the minor cTUs. ATPase complexes translated from the minor cTUs were predicted as the actual F1Fo ATP synthase, because we simultaneously found no or partial A-type ATPase subunits in species with minor cTUs (Suppl. Table 6). Beside, we also identified a batch of chimera fusions consisting of F1Fo ATP synthase subunits. One of these chimera fusions (“β-ε-a-c-b-α-γ”) is identified as part of the N-ATPase operons (Dibrova et al. 2010). In agreement with the previous results on the N-ATPase (Dibrova et al. 2010), we found this chimera fusion transfer between bacterial species as one single unit. Interestingly, the N-ATPase operons share a similar architecture including the “β-ε,” “a-c-b-α-γ,” and one or two additional subunits between the first two modules (Suppl. Fig. 6E). Considering the early division of the N-ATPase, an ancient fusion event possibly participated in the formation of N-ATPase. The presence of these minor cTUs and chimera fusions strongly supports the involvement of distinct modules in the evolution of bacterial F1Fo ATP synthase.
The Subunits in the Central and Peripheral Stalks May Provide the Link for the Integration of F1 and Fo
One of the common features observed by both the phylogenetic profiling and bacterial transcription unit analyses was that OSCP (δ in bacteria) and γ subunits were tightly gathered together with α and β subunits in the F1 component. More specifically, using a variety of independent methods, when the group number was set to 10, phylogenetic profiling results revealed that OSCP and γ subunits were consistently clustered into the same group with the α and β subunits (Fig. 1a, b). Additionally, transcription units such as “ε-β,” “γ-α-δ-b,” “ε-β-γ-α-δ,” and “ε-β-γ-α-δ-b” were frequently detected in bacterial genomes (Fig. 3b). These data, in addition to the known structure of F1Fo ATP synthase, showing direct interaction between γ and OSCP and the α3β3 hexamer (Rubinstein et al. 2003; Carbajo et al. 2005), suggest that these two subunits evolved together with the α3β3 hexamer to form the F1 component. In this model, the ancestral F1 and Fo subunits initially evolved separately (Mulkidjanian et al. 2007); the ancestral α3β3 hexamer, central stalk and part of peripheral stalk then may have formed as a stable protein complex. They were finally integrated with the self-assembled membrane-bound c-subunit ring structure. Bacterial and yeast studies have identified assembly intermediates of the F1Fo ATP synthase. They have not, however, determined whether the lateral stalk forms by sequential attachment of one of its subunits to the c ring or if it exists as an independent unit, which would then incorporate into the Fo (Rak et al. 2011; Houstek et al. 2006; Ruhle and Leister 2015). Our findings of the co-evolution of OSCP (δ in bacteria) with the α3β3 hexamer and the presence of the δ subunit in the same iTUs as α and b subunits suggest sequential attachment of the stalk subunits to the F1 compartment first and then integration into the Fo. The interaction of the b subunit and the stalk subunits with the c-subunit ring, therefore, may provide the link for the integration of F1 and Fo.
Conclusion
Based on our analysis of the phylogenetic profiles of the human mitochondrial proteins, we present the presence of intermediary evolutionary modules for the F-type ATP synthase complex, in addition to the previously known overall F1 and Fo units. The intermediary modules comprise the α3β3 hexamer in the F1 and the c-subunit ring in the Fo as well as the central and lateral stalk subunits of the ATP synthase. Analysis of the bacterial transcription units supported the presence of the additional modules, as highly conserved combinations of separate transcription units are present among certain bacterial classes and phyla. Based on these findings, we propose a model that includes the involvement of multiple modules in the evolution of F1Fo ATP synthase. The central and peripheral stalk subunits provide a link for the integration of the F1/Fo modules.
Abbreviations
- OSCP:
-
Oligomycin sensitivity conferral protein
- HGT:
-
Horizontal gene transfer
- ECMs:
-
Evolutionarily conserved modules
- CLIME:
-
Clustering by inferred models of evolution
- cTU:
-
Conserved transcription unit
- iTU:
-
Incomplete transcription unit
References
Alavian KN, Beutner G, Lazrove E, Sacchetti S, Park HA, Licznerski P, Li H, Nabili P, Hockensmith K, Graham M et al (2014) An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc Natl Acad Sci USA 111(29):10580–10585
Battistuzzi FU, Hedges SB (2009) A major clade of prokaryotes with ancient adaptations to life on land. Mol Biol Evol 26(2):335–343
Beyenbach KW, Wieczorek H (2006) The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol 209(Pt 4):577–589
Bick AG, Calvo SE, Mootha VK (2012) Evolutionary diversity of the mitochondrial calcium uniporter. Science 336(6083):886
Borghese R, Crimi M, Fava L, Melandri BA (1998) The ATP synthase atpHAGDC (F1) operon from Rhodobacter capsulatus. J Bacteriol 180(2):416–421
Boyer PD (1997) The ATP synthase—a splendid molecular machine. Annu Rev Biochem 66:717–749
Buchanan SK, Walker JE (1996) Large-scale chromatographic purification of F1F0-ATPase and complex I from bovine heart mitochondria. Biochem J 318(Pt 1):343–349
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinform 10:421
Carbajo RJ, Kellas FA, Runswick MJ, Montgomery MG, Walker JE, Neuhaus D (2005) Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit. J Mol Biol 351(4):824–838
Collinson IR, Skehel JM, Fearnley IM, Runswick MJ, Walker JE (1996) The F1F0-ATPase complex from bovine heart mitochondria: the molar ratio of the subunits in the stalk region linking the F1 and F0 domains. Biochemistry 35(38):12640–12646
Das A, Ljungdahl LG (2003) Clostridium pasteurianum F1Fo ATP synthase: operon, composition, and some properties. J Bacteriol 185(18):5527–5535
Deckers-Hebestreit G, Altendorf K (1996) The F0F1-type ATP synthases of bacteria: structure and function of the F0 complex. Annu Rev Microbiol 50:791–824
Dibrova DV, Galperin MY, Mulkidjanian AY (2010) Characterization of the N-ATPase, a distinct, laterally transferred Na+-translocating form of the bacterial F-type membrane ATPase. Bioinformatics 26(12):1473–1476
Falk G, Walker JE (1988) DNA sequence of a gene cluster coding for subunits of the F0 membrane sector of ATP synthase in Rhodospirillum rubrum. Support for modular evolution of the F1 and F0 sectors. Biochem J 254(1):109–122
Gay NJ, Walker JE (1981) The atp operon: nucleotide sequence of the promoter and the genes for the membrane proteins, and the delta subunit of Escherichia coli ATP-synthase. Nucleic Acids Res 9(16):3919–3926
Gibbons C, Montgomery MG, Leslie AG, Walker JE (2000) The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol 7(11):1055–1061
Glazko GV, Mushegian AR (2004) Detection of evolutionarily stable fragments of cellular pathways by hierarchical clustering of phyletic patterns. Genome Biol 5(5):R32
Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M (2001) The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409(6820):637–641
Gruber G, Wieczorek H, Harvey WR, Muller V (2001) Structure-function relationships of A-, F- and V-ATPases. J Exp Biol 204(Pt 15):2597–2605
Havlickova V, Kaplanova V, Nuskova H, Drahota Z, Houstek J (2010) Knockdown of F1 epsilon subunit decreases mitochondrial content of ATP synthase and leads to accumulation of subunit c. Biochim Biophys Acta 1797(6–7):1124–1129
Hilario E, Gogarten JP (1998) The prokaryote-to-eukaryote transition reflected in the evolution of the V/F/A-ATPase catalytic and proteolipid subunits. J Mol Evol 46(6):703–715
Houstek J, Pickova A, Vojtiskova A, Mracek T, Pecina P, Jesina P (2006) Mitochondrial diseases and genetic defects of ATP synthase. Biochim Biophys Acta 1757(9–10):1400–1405
Huynen M, Snel B, Lathe W 3rd, Bork P (2000) Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res 10(8):1204–1210
Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T (1989) Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci USA 86(23):9355–9359
Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acid Res 34(Database issue):D354–D357
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Kensche PR, van Noort V, Dutilh BE, Huynen MA (2008) Practical and theoretical advances in predicting the function of a protein by its phylogenetic distribution. J R Soc Interface 5(19):151–170
Korber BT, Farber RM, Wolpert DH, Lapedes AS (1993) Covariation of mutations in the V3 loop of human immunodeficiency virus type 1 envelope protein: an information theoretic analysis. Proc Natl Acad Sci USA 90(15):7176–7180
Koumandou VL, Kossida S (2014) Evolution of the F0F1 ATP synthase complex in light of the patchy distribution of different bioenergetic pathways across prokaryotes. PLoS Comput Biol 10(9):e1003821
Lathe WC 3rd, Snel B, Bork P (2000) Gene context conservation of a higher order than operons. Trends Biochem Sci 25(10):474–479
Lau WC, Baker LA, Rubinstein JL (2008) Cryo-EM structure of the yeast ATP synthase. J Mol Biol 382(5):1256–1264
Lawrence JG, Roth JR (1996) Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143(4):1843–1860
Lee LK, Stewart AG, Donohoe M, Bernal RA, Stock D (2010) The structure of the peripheral stalk of thermus thermophilus H+-ATPase/synthase. Nat Struct Mol Biol 17(3):373–378
Letunic I, Bork P (2011) Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39(Web Server issue):W475–W478
Lewalter K, Muller V (2006) Bioenergetics of archaea: ancient energy conserving mechanisms developed in the early history of life. Biochim Biophys Acta 1757(5–6):437–445
Li Y, Calvo SE, Gutman R, Liu JS, Mootha VK (2014) Expansion of biological pathways based on evolutionary inference. Cell 158(1):213–225
McCarn DF, Whitaker RA, Alam J, Vrba JM, Curtis SE (1988) Genes encoding the alpha, gamma, delta, and four F0 subunits of ATP synthase constitute an operon in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 170(8):3448–3458
Mitchell P (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191:144–148
Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV (2007) Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nat Rev Microbiol 5(11):892–899
Muller V, Gruber G (2003) ATP synthases: structure, function and evolution of unique energy converters. Cell Mol Life Sci 60(3):474–494
Nielsen J, Jorgensen BB, van Meyenburg KV, Hansen FG (1984) The promoters of the atp operon of Escherichia coli K12. Mol Gen Genet 193(1):64–71
Noji H, Yasuda R, Yoshida M, Kinosita K Jr (1997) Direct observation of the rotation of F1-ATPase. Nature 386(6622):299–302
Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK et al (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134(1):112–123
Pasek S, Risler JL, Brezellec P (2006) Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics 22(12):1418–1423
Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO (1999) Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA 96(8):4285–4288
Price MN, Huang KH, Arkin AP, Alm EJ (2005) Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res 15(6):809–819
Price MN, Arkin AP, Alm EJ (2006) The life-cycle of operons. PLoS Genet 2(6):e96
Rak M, Gokova S, Tzagoloff A (2011) Modular assembly of yeast mitochondrial ATP synthase. EMBO J 30(5):920–930
Romero PR, Karp PD (2004) Using functional and organizational information to improve genome-wide computational prediction of transcription units on pathway-genome databases. Bioinformatics 20(5):709–717
Rubinstein JL, Walker JE, Henderson R (2003) Structure of the mitochondrial ATP synthase by electron cryomicroscopy. EMBO J 22(23):6182–6192
Ruhle T, Leister D (2015) Assembly of FF-ATP synthases. Biochim Biophys Acta 1847:849–860
Santana M, Ionescu MS, Vertes A, Longin R, Kunst F, Danchin A, Glaser P (1994) Bacillus subtilis F0F1 ATPase: DNA sequence of the atp operon and characterization of atp mutants. J Bacteriol 176(22):6802–6811
Shih PM, Matzke NJ (2013) Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc Natl Acad Sci USA 110(30):12355–12360
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE (2000) The rotary mechanism of ATP synthase. Curr Opin Struct Biol 10(6):672–679
Sumi M, Yohda M, Koga Y, Yoshida M (1997) F0F1-ATPase genes from an archaebacterium, Methanosarcina barkeri. Biochem Biophys Res Commun 241(2):427–433
Tamames J (2001) Evolution of gene order conservation in prokaryotes. Genome Biol 2(6):RESEARCH0020
Tzagoloff A (1969) Assembly of the mitochondrial membrane system. II. Synthesis of the mitochondrial adenosine triphosphatase. F1. J Biol Chem 244(18):5027–5033
von Ballmoos C, Cook GM, Dimroth P (2008) Unique rotary ATP synthase and its biological diversity. Annu Rev Biophys 37:43–64
Walker JE (1998) ATP synthesis by rotary catalysis (Nobel lecture). Angew Chem Int Ed 37(17):2308–2319
Wang M, Caetano-Anolles G (2009) The evolutionary mechanics of domain organization in proteomes and the rise of modularity in the protein world. Structure 17(1):66–78
Wang H, Oster G (1998) Energy transduction in the F1 motor of ATP synthase. Nature 396(6708):279–282
Xing J, Liao JC, Oster G (2005) Making ATP. Proc Natl Acad Sci USA 102(46):16539–16546
Acknowledgements
YN was supported by the China Scholarship Council. This work was supported by the Imperial College London, Department of Medicine, Division of Brain Sciences funds to KNA.
Funding
YN was supported by the China Scholarship Council. This work was supported by the Imperial College London, Department of Medicine, Division of Brain Sciences funds to KNA.
Author information
Authors and Affiliations
Contributions
KNA and YN: conceived the project, carried out most of the data acquisition and analysis, prepared the figures, and wrote the manuscript text. SM and SS: contributed to data acquisition and analysis. YY and EAJ: contributed intellectually and assisted with manuscript preparation and proofreading. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2017_9819_MOESM1_ESM.pdf
Supple. File 1: Bacterial transcription unit data with improved prediction method. Supplementary material 1 (PDF 392 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Niu, Y., Moghimyfiroozabad, S., Safaie, S. et al. Phylogenetic Profiling of Mitochondrial Proteins and Integration Analysis of Bacterial Transcription Units Suggest Evolution of F1Fo ATP Synthase from Multiple Modules. J Mol Evol 85, 219–233 (2017). https://doi.org/10.1007/s00239-017-9819-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-017-9819-3