Abstract
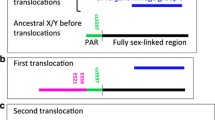
Genes on non-recombining heterogametic sex chromosomes may degrade over time through the irreversible accumulation of deleterious mutations. In papaya, the non-recombining male-specific region of the Y (MSY) consists of two evolutionary strata corresponding to chromosomal inversions occurring approximately 7.0 and 1.9 MYA. The step-wise recombination suppression between the papaya X and Y allows for a temporal examination of the degeneration progress of the young Y chromosome. Comparative evolutionary analyses of 55 X/Y gene pairs showed that Y-linked genes have more unfavorable substitutions than X-linked genes. However, this asymmetric evolutionary pattern is confined to the oldest stratum, and is only observed when recently evolved pseudogenes are included in the analysis, indicating a slow degeneration tempo of the papaya Y chromosome. Population genetic analyses of coding sequence variation of six Y-linked focal loci in the oldest evolutionary stratum detected an excess of nonsynonymous polymorphism and reduced codon bias relative to autosomal loci. However, this pattern was also observed for corresponding X-linked loci. Both the MSY and its corresponding X-specific region are pericentromeric where recombination has been shown to be greatly reduced. Like the MSY region, overall selective efficacy on the X-specific region may be reduced due to the interference of selective forces between highly linked loci, or the Hill–Robertson effect, that is accentuated in regions of low or suppressed recombination. Thus, a pattern of gene decay on the X-specific region may be explained by relaxed purifying selection and widespread genetic hitchhiking due to its pericentromeric location.



Similar content being viewed by others
References
Arguello JR, Zhang Y, Kado T, Fan C, Zhao R, Innan H, Wang W, Long M (2010) Recombination yet inefficient selection along the Drosophila melanogaster subgroup’s fourth chromosome. Mol Biol Evol 27:848–861
Bachtrog D (2004) Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat Genet 36:518–522
Bachtrog D (2008) The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 179:1513–1525
Bachtrog D (2011) Plant sex chromosomes: a non-degenerated Y? Curr Biol 21:R685–R688
Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14:113–124
Bachtrog D, Charlesworth B (2002) Reduced adaptation of a non-recombining neo-Y chromosome. Nature 416:323–326
Bachtrog D, Hom E, Wong KM, Maside X, de Jong P (2008) Genomic degradation of a young Y chromosome in Drosophila miranda. Genome Biol 9:R30
Bachtrog D, Kirkpatrick M, Mank JE, McDaniel SF, Pires JC, Rice WR, Valenzuela N (2011) Are all sex chromosomes created equal? Trends Genet 27:350–357
Bergero R, Charlesworth D (2009) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24:94–102
Bergero R, Charlesworth D (2011) Preservation of the Y transcriptome in a 10-million-year-old plant sex chromosome system. Curr Biol 21:1470–1474
Betancourt AJ, Welch JJ, Charlesworth B (2009) Reduced effectiveness of selection caused by a lack of recombination. Curr Biol 19:655–660
Birky CW Jr, Walsh JB (1988) Effects of linkage on rates of molecular evolution. Proc Natl Acad Sci USA 85:6414–6418
Brown JE, Bauman JM, Lawrie JF, Rocha OJ, Moore RC (2012) The structure of morphological and genetic diversity in natural populations of Carica papaya (Caricaceae) in Costa Rica. Biotropica 44:179–188
Charlesworth B (1996) The evolution of chromosomal sex determination and dosage compensation. Curr Biol 6:149–162
Charlesworth B (2012) The effects of deleterious mutations on evolution at linked sites. Genetics 190:5–22
Charlesworth D (2013) Plant sex chromosome evolution. J Exp Bot 64:405–420
Charlesworth D, Mank JE (2010) The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics 186:9–31
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity 95:118–128
Charlesworth B, Betancourt AJ, Kaiser VB, Gordo I (2009) Genetic recombination and molecular evolution. Cold Spring Harb Symp Quant Biol 74:177–186
Chen CX, Yu QY, Hou SB, Li YJ, Eustice M, Skelton RL, Veatch O, Herdes RE, Diebold L, Saw J, Feng Y, Qian WB, Bynum L, Wang L, Moore PH, Paull RE, Alam M, Ming R (2007) Construction of a sequence-tagged high-density genetic map of papaya for comparative structural and evolutionary genomics in brassicales. Genetics 177:2481–2491
Chibalina MV, Filatov DA (2011) Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr Biol 21:1475–1479
Choo KHA (1998) Why is the centromere so cold? Genome Res 8:81–82
Fay JC, Wu CI (2000) Hitchhiking under positive Darwinian selection. Genetics 155:1405–1413
Gschwend AR, Weingartner LA, Moore RC, Ming R (2012a) The sex-specific region of sex chromosomes in animals and plants. Chromosome Res 20:57–69
Gschwend AR, Yu QY, Tong EJ, Zeng FC, Han J, VanBuren R, Aryal R, Charlesworth D, Moore PH, Paterson AH, Ming R (2012b) Rapid divergence and expansion of the X chromosome in papaya. Proc Natl Acad Sci USA 109:13716–13721
Haddrill PR, Halligan DL, Tomaras D, Charlesworth B (2007) Reduced efficacy of selection in regions of the Drosophila genome that lack crossing over. Genome Biol 8:R18
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program from Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Handel MA, Park C, Kot M (1994) Genetic control of sex-chromosome inactivation during male meiosis. Cytogenet Cell Genet 66:83–88
Hartfield M, Otto SP (2011) Recombination and hitchhiking of deleterious alleles. Evolution 65:2421–2434
Hill WG, Robertson A (1966) The effect of linkage on limits to artificial selection. Genet Res 8:269–294
Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5:R85
Kaiser VB, Charlesworth B (2010) Muller’s ratchet and the degeneration of the Drosophila miranda neo-Y chromosome. Genetics 185:U339–U491
Kirkpatrick M, Guerrero RF (2014) Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics 197:531–541
Kliman RM, Hey J (1993) Reduced natural selection associated with low recombination in Drosophila melanogaster. Mol Biol Evol 10:1239–1258
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Marais G (2003) Biased gene conversion: implications for genome and sex evolution. Trends Genet 19:330–338
Marais GAB, Nicolas M, Bergero R, Chambrier P, Kejnovsky E, Moneger F, Hobza R, Widmer A, Charlesworth D (2008) Evidence for degeneration of the Y chromosome in the dioecious plant Silene latifolia. Curr Biol 18:545–549
McVean GAT, Charlesworth B (2000) The effects of Hill-Robertson interference between weakly selected mutations on patterns of molecular evolution and variation. Genetics 155:929–944
Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA (2002) Spermatozoal RNA profiles of normal fertile men. Lancet 360:772–777
Rand DM, Kann LM (1996) Excess amino acid polymorphism in mitochondrial DNA: contrasts among genes from Drosophila, mice, and humans. Mol Biol Evol 13:735–748
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Method Mol Biol 132:365–386
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tajima F (1993) Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135:599–607
Wang JP, Na JK, Yu QY, Gschwend AR, Han J, Zeng FC, Aryal R, VanBuren R, Murray JE, Zhang WL, Navajas-Perez R, Feltus FA, Lemke C, Tong EJ, Chen CX, Wai CM, Singh R, Wang ML, Min XJ, Alam M, Charlesworth D, Moore PH, Jiang JM, Paterson AH, Ming R (2012) Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci USA 109:13710–13715
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Weingartner LA, Moore RC (2012) Contrasting patterns of X/Y polymorphism distinguish Carica papaya from other sex chromosome systems. Mol Biol Evol 29:3909–3920
Wright SI, Charlesworth B (2004) The HKA test revisited: a maximum-likelihood-ratio test of the standard neutral model. Genetics 168:1071–1076
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13:555–556
Yang ZH (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yu QY, Hou SB, Hobza R, Feltus FA, Wang X, Jin WW, Skelton RL, Blas A, Lemke C, Saw JH, Moore PH, Alam M, Jiang JM, Paterson AH, Vyskot B, Ming R (2007) Chromosomal location and gene paucity of the male specific region on papaya Y chromosome. Mol Genet Genomics 278:177–185
Yu QY, Hou S, Feltus FA, Jones MR, Murray JE, Veatch O, Lemke C, Saw JH, Moore RC, Thimmapuram J, Liu L, Moore PH, Alam M, Jiang JM, Paterson AH, Ming R (2008a) Low X/Y divergence in four pairs of papaya sex-linked genes. Plant J 53:124–132
Yu QY, Navajas-Perez R, Tong E, Robertson J, Moore PH, Paterson AH, Ming R (2008b) Recent origin of dioecious and gynodioecious chromosomes in papaya. Tropical Plant Biol 1:49–57
Yu QY, Tong E, Skelton RL, Bowers JE, Jones MR, Murray JE, Hou SB, Guan PZ, Acob RA, Luo MC, Moore PH, Alam M, Paterson AH, Ming R (2009) A physical map of the papaya genome with integrated genetic map and genome sequence. BMC Genom 10:371–382
Zeng K, Shi S, Wut CI (2007) Compound tests for the detection of hitchhiking under positive selection. Mol Biol Evol 24:1898–1908
Zhang WL, Wang XU, Yu QY, Ming R, Jiang JM (2008) DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res 18:1938–1943
Zhou Q, Bachtrog D (2012a) Chromosome-wide gene silencing initiates Y degeneration in Drosophila. Curr Biol 22:522–525
Zhou Q, Bachtrog D (2012b) Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science 337:341–345
Acknowledgments
We thank two anonymous reviewers for their helpful comments in preparing this manuscript. This work was supported by the National Science Foundation (DBI-0922545) to RCM and by the Department of Botany at Miami University through Academic Challenge grants to MW. We are greatly indebted to Oscar Rocha at Kent State University for his help during the specimen collecting process in Costa Rica. We would also like to thank our collaborators Ray Ming (University of Illinois at Urbana-Champaign) and Qingyi Yu (Texas A&M University) for providing papaya sequence information and cultivar tissues.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, M., Moore, R.C. The Evolutionary Tempo of Sex Chromosome Degradation in Carica papaya . J Mol Evol 80, 265–277 (2015). https://doi.org/10.1007/s00239-015-9680-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-015-9680-1