Abstract
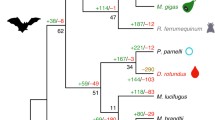
Frugivorous and nectarivorous bats rely largely on hepatic glycogenesis and glycogenolysis for postprandial blood glucose disposal and maintenance of glucose homeostasis during short time starvation, respectively. The glycogen synthase 2 encoded by the Gys2 gene plays a critical role in liver glycogen synthesis. To test whether the Gys2 gene has undergone adaptive evolution in bats with carbohydrate-rich diets in relation to their insect-eating sister taxa, we sequenced the coding region of the Gys2 gene in a number of bat species, including three Old World fruit bats (OWFBs) (Pteropodidae) and two New World fruit bats (NWFBs) (Phyllostomidae). Our results showed that the Gys2 coding sequences are highly conserved across all bat species we examined, and no evidence of positive selection was detected in the ancestral branches leading to OWFBs and NWFBs. Our explicit convergence test showed that posterior probabilities of convergence between several branches of OWFBs, and the NWFBs were markedly higher than that of divergence. Three parallel amino acid substitutions (Q72H, K371Q, and E666D) were detected among branches of OWFBs and NWFBs. Tests for parallel evolution showed that two parallel substitutions (Q72H and E666D) were driven by natural selection, while the K371Q was more likely to be fixed randomly. Thus, our results suggested that the Gys2 gene has undergone parallel evolution on amino acid level between OWFBs and NWFBs in relation to their carbohydrate metabolism.


Similar content being viewed by others
References
Abascal F, Zardoya R, Posada D (2005) ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105
Altingham JD, McOwat T, Hammond L (1998) Bats: biology and behavior. Oxford University Press, Oxford
Anisimova M, Bielawski JP, Yang Z (2002) Accuracy and power of Bayes prediction of amino acid sites under positive selection. Mol Biol Evol 19:950–958
Barclay RMR, Barclay LE, Jacobs DS (2006) Deliberate insectivory by the fruit bat Rousettus aegyptiacus. Acta Chiropt 8:549–553
Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions. Bioinformatics for geneticists. Wiley, New York
Bielawski JP, Yang Z (2003) Maximum likelihood methods for detecting adaptive evolution after gene duplication. J Struct Funct Genomics 3:201–212
Bollen M, Keppens S, Stalmans W (1998) Specific features of glycogen metabolism in the liver. Biochem J 336:19–31
Browner MF, Nakano K, Bang AG, Fletterick RJ (1989) Human muscle glycogen synthase cDNA sequence: a negatively charged protein with an asymmetric charge distribution. Proc Natl Acad Sci USA 86:1443–1447
Brunner Y, Schvartz D, Priego-Capote F, Couté Y, Sanchez JC (2009) Glucotoxicity and pancreatic proteomics. J Proteomic 71:576–591
Capaldo B, Gastaldelli A, Antoniello S, Auletta M, Pardo F, Ciociaro D, Guida R, Ferrannini E, Saccà L (1999) Splanchnic and leg substrate exchange after ingestion of a natural mixed meal in humans. Diabetes 48:958–966
Castoe TA, de Koning AP, Kim HM, Gu W, Noonan BP, Naylor G, Jiang ZJ, Parkinson CL, Pollock DD (2009) Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci USA 106:8986–8991
Caviedes-Vidal E, McWhorter TJ, Lavin SR, Chediack JG, Tracy CR, Karasov WH (2007) The digestive adaptation of flying vertebrates: high intestinal paracellular absorption compensates for smaller guts. Proc Natl Acad Sci USA 104:19132–19137
Caviedes-Vidal E, Karasov WH, Chediack JG, Fasulo V, Cruz-Neto AP, Otani L (2008) Paracellular absorption: a bat breaks the mammal paradigm. PLoS One 3:e1425
Consoli A, Kennedy F, Miles J, Gerich J (1987) Determination of Krebs cycle metabolic carbon exchange in vivo and its use to estimate the individual contributions of gluconeogenesis and glycogenolysis to overall glucose output in man. J Clin Invest 80:1303–1310
Datzmann T, von Helversen O, Mayer F (2010) Evolution of nectarivory in phyllostomid bats (Phyllostomidae Gray, 1825, Chiroptera: Mammalia). BMC Evol Biol 10:165
Gitzelmann R, Spycher MA, Feil G, Müller J, Seilnacht B, Stahl M, Bosshard NU (1996) Liver glycogen synthase deficiency: a rarely diagnosed entity. Eur J Pediatr 155:561–567
Herrera MLG, Hobson KA, Mirón ML, Ramírez PN, Méndez CG, Sánchez-Cordero V (2001) Sources of protein in two species of phytophagous bats in a seasonal dry forest: evidence from stable-isotope analysis. J Mamm 82:352–361
Herrera GL, Gutierrez E, Hobson KA, Altube B, Díaz WG, Sánchez-Cordero V (2002) Sources of assimilated protein in five species of New World frugivorous bats. Oecologia 133:280–287
Huang S, Czech MP (2007) The GLUT4 glucose transporter. Cell Metab 5:237–252
Irimia JM, Meyer CM, Peper CL, Zhai L, Bock CB, Previs SF, McGuinness OP, DePaoli-Roach A, Roach PJ (2010) Impaired glucose tolerance and predisposition to the fasted state in liver glycogen synthase knock-out mice. J Biol Chem 285:12851–12861
Kawahito S, Kitahata H, Oshita S (2009) Problems associated with glucose toxicity: role of hyperglycemia-induced oxidative stress. World J Gastroenterol 15:4137–4142
Keegan DJ (1977) Aspects of the assimilation of sugars by Rousettus aegyptiacus. Comp Biochem Physiol A Physiol 58:349–352
Klip A (2009) The many ways to regulate glucose transporter 4. Appl Physiol Nutr Metab 34:481–487
Laska M (1990) Food transit times and carbohydrate use in three phyllostomid bat species. Z Säugetierkunde 55:49–54
Liu Y, Han N, Franchini LF, Xu H, Pisciottano F, Elgoyhen AB, Rajan KE, Zhang S (2011a) The voltage-gated potassium channel subfamily KQT member 4 (KCNQ4) displays parallel evolution in echolocating bats. Mol Biol Evol 29:1441–1450
Liu Z, Li S, Wang W, Xu D, Murphy RW, Shi P (2011b) Parallel evolution of KCNQ4 in echolocating bats. PLoS One 6:e26618
Ludvik B, Nolan JJ, Roberts A, Baloga J, Joyce M, Bell JM, Olefsky JM (1995) A noninvasive method to measure splanchnic glucose uptake after oral glucose administration. J Clin Invest 95:2232–2238
Nuttall FQ, Gannon MC, Bai G, Lee EY (1994) Primary structure of human liver glycogen synthase deduced by cDNA cloning. Arch Biochem Biophys 311:443–449
Orho M, Bosshard NU, Buist NR, Gitzelmann R, Aynsley-Green A, Blümel P, Gannon MC, Nuttall FQ, Groop LC (1998) Mutations in the liver glycogen synthase gene in children with hypoglycemia due to glycogen storage disease type 0. J Clin Invest 102:507–515
Parry-Jones K, Augee ML (1992) Insects in flying fox diets. Bat Res News 33:9–11
Pederson BA, Cheng C, Wilson WA, Roach PJ (2000) Regulation of glycogen synthase. Identification of residues involved in regulation by the allosteric ligand glucose-6-P and by phosphorylation. J Biol Chem 275:27753–27761
Pinheiro EC, Taddei VA, Migliorini RH, Kettelhut IC (2006) Effect of fasting on carbohydrate metabolism in frugivorous bats (Artibeus lituratus and Artibeus jamaicensis). Comp Biochem Physiol B 143:279–284
Pond SL, Frost SD, Muse SV (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679
Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS (2012) Glycogen and its metabolism: some new developments and old themes. Biochem J 441:763–787
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Rothman DL, Magnusson I, Katz LD, Shulman RG, Shulman GI (1991) Quantitation of hepatic glycogenolysis and gluconeogenesis in fasting humans with 13C NMR. Science 254:573–576
Shen YY, Liu J, Irwin DM, Zhang YP (2010) Parallel and convergent evolution of the dim-light vision gene RH1 in bats (Order: Chiroptera). PLoS One 5:e8838
Shen B, Han X, Zhang J, Rossiter SJ, Zhang S (2012) Adaptive evolution in the glucose transporter 4 gene Slc2a4 in Old World fruit bats (Family: Pteropodidae). PLoS One 7:e33197
Shepherd PR, Kahn BB (1999) Glucose transporters and insulin action—implications for insulin resistance and diabetes mellitus. N Engl J Med 341:248–257
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Taylor R, Magnusson I, Rothman DL, Cline GW, Caumo A, Cobelli C, Shulman GI (1996) Direct assessment of liver glycogen storage by 13C nuclear magnetic resonance spectroscopy and regulation of glucose homeostasis after a mixed meal in normal subjects. J Clin Invest 97:126–132
Teeling EC, Springer MS, Madsen O, Bates P, O’Brien SJ, Murphy WJ (2005) A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Thorens B, Mueckler M (2010) Glucose transporters in the 21st century. Am J Physiol Endocrinol Metab 298:E141–E145
Tracy CR, McWhorter TJ, Korine C, Wojciechowski MS, Pinshow B, Karasov WH (2007) Absorption of sugars in the Egyptian fruit bat (Rousettus aegyptiacus): a paradox explained. J Exp Biol 210:1726–1734
Voigt CC, Zubaid A, Kunz TH, Kingston T (2011) Sources of assimilated proteins in Old and New World phytophagous bats. Biotropica 43:108–113
Weinstein DA, Correia CE, Saunders AC, Wolfsdorf JI (2006) Hepatic glycogen synthase deficiency: an infrequently recognized cause of ketotic hypoglycemia. Mol Genet Metab 87:284–288
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Yang Z, Bielawski JP (2000) Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503
Yang Z, Kumar S, Nei M (1995) A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141:1641–1650
Zhang J, Kumar S (1997) Detection of convergent and parallel evolution at the amino acid sequence level. Mol Biol Evol 14:527–536
Zhang J, Nielsen R, Yang Z (2005) Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22:2472–2479
Zhou X, Xu S, Xu J, Chen B, Zhou K, Yang G (2012) Phylogenomic analysis resolves the interordinal relationships and rapid diversification of the Laurasiatherian mammals. Syst Biol 61:150–164
Acknowledgments
The authors thank Yi-Hsuan Pan for technical advice on protein analyses and Ming Lei for help in convergent evolution analyses. This study was supported by the Foundation for Science and Technology Innovation of University Students from East China Normal University to YQ, the Chinese National Science Foundation (Grant No. 31172077) to SZ, and the Academic Scholarship for Ph.D. Candidate Granted by Ministry of Education of the P.R.China (MXRZZ2012008) to BS.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Schematic representation of biochemical pathway of glycogen synthesis. Blood glucose is transported into cells via glucose transporter proteins and converted into glucose 6-phosphate by hexokinase 4. Then glucose 6-phosphate is converted into glucose 1-phosphate by phosphoglucomutase. And the glucose 1-phosphate is converted into UDP-glucose by UDP-glucose pyrophosphorylase. Finally, the UDP-glucose is used as glucosyl donor by glycogen synthase to form glycogen via formation of the α-1,4-glycosidic linkages (PDF 289 kb)
Fig. S2
Alignment of the amino acid sequences of the Gys2 gene from 22 mammals (only the variable sites are shown) (PDF 374 kb)
Fig. S3
Nonsynonymous amino acid substitutions mapped onto the species topology of 22 mammals. Branch lengths are not drawn to scale (PDF 365 kb)
Fig. S4
Species topology showing inferred evolutionary change along the Gys2 gene. Values on each branch are rates of non-synonymous substitution (d N) and synonymous substitution (d S) estimated by one-ratio model based on maximum-likelihood method using PAML CODEML package. Branch lengths are based on nucleotide substitutions per codon (PDF 312 kb)
Rights and permissions
About this article
Cite this article
Qian, Y., Fang, T., Shen, B. et al. The Glycogen Synthase 2 Gene (Gys2) Displays Parallel Evolution Between Old World and New World Fruit Bats. J Mol Evol 78, 66–74 (2014). https://doi.org/10.1007/s00239-013-9600-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-013-9600-1