Abstract
Protochordate genomes enable a prevalence of hemostasis evolution. Broad searches were performed for homologs of human serine proteases of hemostasis on the genomes of Branchiostoma floridae, Saccoglossus kowalevskii, and Strongylocentrotus purpuratus. Sequences were analyzed by multiple bioinformatic tools. The survey revealed numerous homologous components. Amphioxus was rich in some serine proteases not accompanied by gamma-carboxyglutamic or kringle domains similar more to thrombin than to other coagulation factors. The serine proteases found in amphioxus exhibited the attributes similar to those of thrombin by phylogeny relationships, sequence conservation, gene synteny, spatial structure, and ligand docking. A few plasminogen- and plasminogen activators-like proteases with kringles were also present. Those serine proteases demonstrated the greatest proximity rather to plasminogen or plasminogen activators than to thrombin. Searching for homologs of serine protease hemostatic factors in acorn worm and sea urchin revealed several components similar to those found in amphioxus. Hypothetically, the common ancestor of chordates had three separate serine proteases that evolved independently into immunoglobulin-like and kringle proteases in lancelets, and prothrombin, plasminogen activators, and plasminogen in vertebrates. Ancestral proteases evolved in vertebrates into hemostasis factors after merging the proper N-terminal domains and duplications.
Similar content being viewed by others
Introduction
Molecular cloning and availability of the whole genome sequences of chordates enables reconstruction of complex processes like hemostasis (Davidson et al. 2003a; Davidson et al. 2003b; Doolittle 1993; Doolittle et al. 2008; Doolittle 2009, 2011, 2012; Doolittle and Feng 1987; Jiang and Doolittle 2003; Escriva et al. 2002; Loof et al. 2011), complement system (Nonaka and Yoshizaki 2004; Suzuki et al. 2002) and embryonic development (Krem and Di Cera 2002). It has been reported that lampreys have simpler coagulation system with sole predecessor genes corresponding to human blood coagulation factors IX (FIX) and X (FX) and their cofactors: factor V (FV) and VIII (FVIII) (Doolittle et al. 2008; Kimura et al. 2009). On the other hand the intrinsic phase of clotting is much younger and appears to be characteristic only for placental and marsupial mammals. Monotremes, birds, and amphibians have lone predecessor gene for factor XI/plasma prekallikrein (FXI/PK) but fish lack the genes for proteases of the contact phase of blood coagulation (Table 1) (Ponczek et al. 2008).
Recently performed whole genome sequencing of exponents of primitive chordates like amphioxus Branchiostoma floridae (Putnam et al. 2008) and sea squirt Ciona Intestinalis (Dehal et al. 2002), hemichordates like acorn worm Saccoglossus kowalevskii and echinoderms such as sea urchin Strongylocentrotus purpuratus (Sodergren et al. 2006) enables moving back before divergence of lamprey in analysis of evolution of hemostasis.
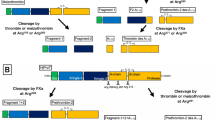
The chain of cascade reactions involving several serine proteases from chymotrypsin subfamily in the form of zymogens converted to active enzymes shapes the hemostatic system of vertebrates (Riddel et al. 2007; Schenone et al. 2004). These proteases are blood coagulation factors VII (FVII), FIX, FX, XII (FXII), FXI, PK, protein C (PC), protein Z and the key protein of coagulation: prothrombin (T), as well as proteases of fibrinolysis like plasminogen (Plg) and two plasminogen activators—tissue (tPA) and urokinase (uPA) (Fig. 1a).
Hemostasis and hemostatic proteases (Riddel et al. 2007; Schenone et al. 2004). a Schematic presentation of mammalian hemostasis (right); hypothetical hemostasis of lamprey (left); F coagulation factors, FBG fibrinogen, FBN fibrin, FBX fibrin stabilized by factor XIII, FDP fibrin degradation products, FDPX stabilized fibrin degradation products; KNG high molecular kininogen, PK plasma prekallikrein, PL phospholipids, PLG plasminogen, PLN plasmin, tPA tissue plasminogen activator, uPA urokinase-type plasminogen activator, solid lines coagulation, dotted lines fibrinolysis. b Domain structures of prothrombin, plasminogen, tissue plasminogen activator, urokinase plasminogen activator, and factors VII, IX, and X, XI, XII. T protease domain, P PAN domain, K kringle domain, F fibronectin domain, E EGF domain (based on Doolittle et al. 2008; Doolittle 2009, 2011 and protein sequence PROSITE anticipation—http://www.expasy.ch/prosite)
The proteases of extrinsic pathway of blood coagulation—FVII, FX, and FIX share resemblance in sequence and domain arrangement in all vertebrates (Fig. 1b).
The studies of the jawless vertebrates like lamprey, strengthened the arguments about importance of gene duplications in the evolution of hemostasis. The duplications in common ancestor of jawed vertebrates must have resulted in divergence of separate FIX and FX as well as their cofactors (Doolittle et al. 2008). Similarly, analysis of genomes of amphioxus and other primitive chordates and “pre-chordates” might provide information about the beginnings of serine proteases of coagulation system as well as some other factors in predecessors of vertebrates. Sequence similarity and phylogenetic trees based on sequence alignments, occurrence and arrangement of domains, synteny (the co-localization of sets of genes on chromosomes), and spatial structure might serve to shed some additional light on the pre-vertebrate evolution of serine proteases of blood coagulation, with particular emphasis on thrombin. The objective of this study is to answer the questions about relics of serine proteases, related to protease factors of vertebrate hemostasis, present in primitive non-vertebrate deuterostomes. What kinship do they share with vertebrate factors? Have any signs of thrombin-fibrinogen-like mechanisms survived at all? When and how did appear predecessors of modern vertebrate hemostatic proteases?
Methods
Human and the other known vertebrate protein sequences were found on UniProt or NCBI GenBank to be used as templates. Localizations of protein domains were estimated by PROSITE (http://www.expasy.ch/prosite/) (de Castro et al. 2006) and PFAM (http://pfam.sanger.ac.uk/ ). DNA sequences of human chromosomes were downloaded from ENSEMBL Genome Browser (http://www.ensembl.org/). Publicly accessible sequences of B. floridae were downloaded from DOE Joint Genome Institute (JGI) web site: http://genome.jgi-psf.org/. Sequences of Saccoglossus kowalevskii and Strongylocentrotus purpuratus were downloaded from Baylor College of Medicine Human Genome Sequencing Center web site: http://www.hgsc.bcm.tmc.edu/. The Basic Local Alignment Search Tool (BLAST) executables were downloaded from NCBI (Altschul et al. 1997). TBLASTN searches with domains of serine proteases of hemostasis (serine protease and GLA domains of prothrombin, FX, FIX, FVII and protein C; kringles of plasminogen, tPA and uPA) were performed against B. floridae v1.0 unmasked assembly (Bfv1UA) with local BLAST software as well as JGI genome browser. The highest hits were verified by B. floridae v2.0 assembly (Bfv2A) and next blasted against the NCBI non-redundant and nucleotide database. NCBI protein BLAST was used to search for proteins in genome databases of B. floridae, S. purpuratus and S. kowalevskii. Reconstructions between exons were made with GeneScan (Burge and Karlin 1997) and by manual assembles of multiple amino acid sequence alignments and BLAST results. Multiple alignments were made with ClustalW (Thompson et al. 1994). Phylip 3.69 software package was employed to count consensus tree by parsimony method after 100 bootstrap replications (Felsenstein 1989). Mrbayes 3.1.2 (Huelsenbeck and Ronquist 2001) was used to count Markov chain Monte Carlo tree with Blosum62 model (Henikoff and Henikoff 1992). The phylogenetic trees were drawn with iTOL (Letunic and Bork 2007). Serine protease sequence of sea anemone Nematostella vectensis (XP_001633098)* was used as out-group. Synteny analyses and additional DNA comparisons were made with ACT DNA Sequence Comparison Viewer (Carver et al. 2005). Ancestral sequences were reconstructed using ANCESCON (Cai et al. 2004) on the web site http://prodata.swmed.edu/ancescon/ancescon.php.
One predicted amphioxus protease together with its acorn worm homolog and two hypothetical ancestors generated by ANCESCON were chosen for homology modeling with SwissModel in automated mode (Kiefer et al. 2009) and the spatial structures were visualized by Autodock tools (Sotriffer et al. 2000). Ligand docking of three physiological ligands of thrombin with chosen lancelet serine protease was performed using Autodock Vina 1.0 (Trott and Olson 2010).
Homology modeling was performed with 1FPH thrombin structure (Stubbs et al. 1992) as a template. The predicted structure of amphioxus protein was additionally tested by Autodock Vina 1.0 (Trott and Olson 2010) and visualized by Autodock Tools v 1.5.6rc1 (Sotriffer et al. 2000) for binding of 3 peptide ligands typical for thrombin of vertebrates: fragment of fibrinopeptide A: DFLAEGGGVR, fragment of hirudin: DFEEIPEEpY and N-terminal fragment of seven-transmembrane-domain protease activeted receptor (peptide NRS, a part of PAR-1): LDPRSFLLRNPNDKYEPFW 1 (ATNATLDPRSFLLRNPNDKYEPFWE2 ). Structures of fibrinopeptide A and hirudin were extracted from 1FPH and a sequence of PAR1 extracellular fragment (peptide NRS) was taken from 1NRN1 (Mathews et al. 1994) and 3LU92 (Gandhi et al. 2010) using Swiss-PdbViewer DeepView v4.04 http://www.expasy.org/spdbv/ (Guex and Peitsch 1997). The structures of bolded, presented above, sequences were saved as separate PDB files and after conversion to PDBQT files in Autodock Tools used as ligands in Vina.
Results
Searching for Blood Coagulation-Like Serine Proteases
Human amino acid sequences from serine protease domain (containing catalytic triad His57, Asp102, Ser195) of prothrombin, protein C, and factors VII, IX, and X were taken as templates to search for hypothetical orthologs. The best hits were blasted against the NCBI protein BLAST with a selection of optional vertebrate organism, including taxa of Homo, Mus, Gallus, Xenopus, Danio, Branchiostoma, Saccoglossus and Strongylocentrotus. The best result for prothrombin was found on scaffold 396 (Bf_V2_279) of amphioxus genome. The reconstructed protease domain was the first on the list as thrombin, 42, 43, and 43 % identical, respectively, in human, mouse, and frog NCBI back-search. Moreover, the sequence could be identified as hypothetical protein BRAFLDRAFT_241477 (alternative nomination XP_002587250 or EEN43261)** when back-searched against B. floridae. No GLA, kringle or EGF domains could be found in the upstream region of the gene. Surprisingly, an exon for immunoglobulin-like (IG-like) domain between hypothetic promoter and serine protease domain was found. The sequence was flanked by introns with AG and GT, respectively. Blasting of IG-like and serine protease sequences with no species selection resulted in 6 hits of B. floridae homologs with similarity varying from 49 to 73 % (including BRAFLDRAFT_241477 or XP_002587250 100 % hit). The seventh hit was 34 % identical protein described as “similar to Low-density lipoprotein-receptor-related protein 4 precursor” from sea urchin Strongylocentrotus purpuratus, (XP_783851)*** and the eighth serine protease was putative-like of Saccoglossus kowalevskii (XP_002740278)****. The sea urchin protein appeared to be a serine protease with 4 low-density lipoprotein-receptor domains, EGF domain, 2 sushi repeats, and 2 IG-like domains. The acorn worm protease had no additional domains. The protease domains of sea urchin and acorn worm were 41 and 45 % identical with lancelet protein, respectively. Protease domain of human prothrombin blasted against the sea urchin and acorn worm genomes returned the same proteins (XP_783851 43 % identical and XP_002740278 42 % identical). The next hits were similar below 40 % both for echinoderms and hemichordates. Acorn worm had one more paralog XP_002737308, 47 % identical with XP_002740278 but that additional protein had C-terminal FReD domain. The back-search against human gave 40 % identical prothrombin for protease domain a template and 42 % identical fibrinogen-like-1 for FReD domain as template. Searching the amphioxus serine protease domain against echinoderms revealed additional 40 % identical serine protease with IG-like domain (similar to Lpa-prov protein (XP_788297.2) in the sea urchin genome. The next hits, 35 and 38 % identical respectively, were described as “similar to thrombin” (XP_001193031.1) and “similar to prothrombin precursor” (XP_001176043.1). The first record had IG-like domain, but the second was equipped with 3 fibronectin domains and one SRCR domain.
Alignments of the lancelet scaffold 396 protein with vertebrate prothrombins showed that amphioxus protein had 100 % conserved motif DACEGD S GGPF within serine protease with serine residue from catalytic triad almost in the middle (underlined) and 66 % conserved sequence VSWG D GC AL (differences with human in italics). Their equivalents are responsible for the binding of fibrinopeptide A in vertebrate thrombin. Moreover two more proteins were found with the same conserved motifs in B. floridae. The first denominated in databases as XP_002593216, EEN49227.1 or BRAFLDRAFT_209883 was 75 % identical with the scaffold 396 protein and the second—XP_002601488 (EEN57500, BRAFLDRAFT_241809) was 70 % identical. Sequences characteristic for hirudin and protease-activated receptor (PAR) binding (peptide NRS), which are located in thrombin exosite I, were not very well conserved, but Exosite II (human following sequences: Arg93-Ile103: RYNWRENLDRDI, Arg185-Ser195: RGDACEGDS, Val213-Gly226: VSWGEGCDRDGKYG) (Mathews et al. 1994) and Na+ binding site (human following sequences: Met180-Tyr184: MFCAGY, Lys224-Tyr228: KYGFY, Val213-Gly219: VSWGEG, Gly188-Glu192: GDACE) were conservative (Fig. 2) (Di Cera 2003; Di Cera 2007; Di Cera 2008; Di Cera 2009; Di Cera et al. 1995). The close relation of scaffold 396 (XP_002587250) protein and the other amphioxus, acorn worm and sea urchin proteases with prothrombin of vertebrates was confirmed both by a phylogenetic tree calculated with bootstrapping and by Markov chain Monte Carlo method. The thrombin-like proteases of amphioxus**, acorn worm****, and sea urchin*** form a common clade with vertebrate prothrombin serine protease domain (Fig. 3).
Phylogenetic trees generated from the serine protease portions of vertebrate coagulation factors, non-hemostatic proteins and amphioxus, acorn worm, and sea urchin thrombin-like serine proteases. a The parsimony tree calculated after 100 bootstrap replications from an alignment of complete serine protease regions. Software used Phylip 3.69 and iTOL. b Markov chain Monte Carlo tree calculated with Blosum62 model. Software used Mrbayes-3.1.2 and iTOL. Proteases: F7 factor VII, F9 factor IX, F10 factor X, HGF hepatocyte growth factor, MASP 1 and 2 mannan-binding lectin serine protease 1 and 2, PC protein C, PT prothrombin, PZ protein Z, PLG plasminogen, TPSB1 anti-tryptase beta-1, TPSG1 tryptase gamma, TPS11B transmembrane protease serine 11B, TR trypsin, TSP50 probable threonine protease PRSS50, PRSS33 probable threonine protease 33, PRSS22 probable threonine protease 22, ACE86411 “plasminogen” of B. belcheri tsingtauense, AAO12215 “trypsin” of sponge Aplysina fistularis, numeral designations B. floridae serine proteases found on NCBI data base (accessions numerals without XP prefix, species Table 3), sca 384, sca 542 serine proteases found in B. floridae genome but absent in NCBI database. HU (and if not mentioned) human (Homo sapiens), MO mouse (Mus musculus), OP opossum (Monodelphis domestica), CH chicken (Gallus gallus), ST ostrich (Struthio camelus), TA zebra finch (Taeniopygia guttata), CR crocodile (Crocodylus niloticus), TU turtle (Trachemys scripta elegans), SN snake (Elaphe sp.), DA zebrafish (Danio reiro), LA lamprey (Lethenteron japonicum), HF hagfish (Eptatretus stoutii),. (Supporting information in Appendix 1 and 2). Putative time of divergence assigned according to Peterson et al. 2008 (&—divergence of all vertebrates, &&—divergence of jawless fish and &&&—divergence of amphioxus)
ACT synteny analysis revealed extensive conservation between amphioxus scaffold 396 and human chromosome 11 (the location of human prothrombin gene). As an example solitary gene for carnitine palmitoyltransferase (in human three genes: CPT1A—Chr. 11, CPT1B—Chr. 22, CPT2C—Chr. 19) could be found for amphioxus and it was placed on scaffold 396 (Fig. 4).
Synteny of amphioxus scaffold 396 and human chromosome 11 (double-stranded DNA). CPT1A carnitine palmitoyltransferase 1A (liver isoform), CPT carnitine palmitoyltransferase-like (only one gene in lancelet), F2 prothrombin, F2L prothrombin-like (XP_002587250). Software used ACT DNA Sequence Comparison Viewer
Serine protease domains of factors VII, IX, X and PC returned many hits against the B. floridae, S. kowalevskii, and S. purpuratus genomes. All homologs were identical less than 41 %, they had no typical amino terminus domains and the back-search pointed on human trypsin identical from 39 to 53 % for lancelet, on human thrombin identical 42 % for acorn worm and on human transmembrane proteases identical 40 % for sea urchin. None of them can be regarded as true orthologs of vertebrate coagulation factors VII, IX, X and PC (Table 1).
Four plasminogen-like proteases (XP_002596007, XP_002590082, XP_002590083, and XP_002596021) were detected in B. floridae using human plasminogen serine protease domain with 5 kringles as template. Amphioxus proteins had only 2 kringle domains instead of 5 like in vertebrate plasminogens. The back-search against human returned plasminogen for all of them with 44, 44, 40, and 34 % identity, respectively. Human hepatocyte growth factor (HGF) (4 kringle paralog of plasminogen) pointed on the same results but with lower similarity (33, 36, 35 and 29 % respectively). Moreover, human plasminogen and HGF kringles returned XP_002609225 in lancelet as a best hit. The record appeared to be 12 kringle protein with transmembrane motif but without serine protease domain. It was 43 and 48 % identical to plasminogen and HGF kringles, respectively. Three additional kringle proteases (XP_002601959, XP_002596022, XP_002596023, and XP_002596018) were found using whole tissue and urokinase-type plasminogen activators as templates. The first sequence had 2 kringle domains and was 41 % similar to HGF. The rest had only one kringle domain. Their protease domains were 46 and 42 % similar to transmembrane protease serine 11F and 44 % similar to hepsin in back-search, respectively. They were only 40–41 % similar to plasminogen and less than 40 % similar to uPA or tPA. When uPA and tPA protease domains were used as templates the best hit in amphioxus was XP_002605710, the protein without kringle domains. It was 40 % similar to both templates. Additional hits were serine proteases that had kringle, SRCR (Scavenger receptor cysteine-rich domain) and LDL domains (LDL receptor domain class). Acorn worm returned record XP_002736981 with protease domains 47, 43, 46, and 45 % identical with XP_002596007, XP_002590082, XP_002590083, and XP_002596021, respectively, but without kringle domains. Additional proteases had mostly CUB domains. Finally, no kringles attached to serine proteases could be found in acorn worm. Similarly, in sea urchin no kringles were associated directly with serine protease domain, however one containing kringles serine protease was found. The protein was described as similar to GRAAL2 protein (with two numbers—XP_001192777 and XP_783458). The protease domain shared 45 % similarity with human apolipoprotein, 42 % with plasminogen, and 40 % with chymotrypsinogen B2 protease domains. The protein appeared to be a real mosaic. It had SRCR, kringle, two PANs, kringle, discoidin, LDL, PAN, LDL and SRCR before C-terminus of serine protease domain.
GLA Domains
GLA sequences from human prothrombin, FX, FIX, FVII, and protein C to search the sea urchin, acorn worm and amphioxus genomes were used. No GLA domains were found in sea urchin whereas only one GLA domain was found in acorn worm. It was located at N-terminus of a multiple EGF protein (XP_002740003) and was 54 % similar to human GLA of prothrombin. Two GLA domains were found to be encoded in the lancelet genome. The first GLA was located in a protein XP_002599334 found on scaffold 4 (Bfv1UA, Bfv2A: Bf_V2_145), and it was 45 % identical with GLA of human prothrombin. Blasting of GLA sequence against NCBI returned GLA domains of proline-rich Gla protein and jawed vertebrate coagulation factors IX, X, VII, protein S, protein C, prothrombin, as well as FVII, FX, and prothrombin in lamprey. The second additional, incomplete GLA protein (XP_002599944) was found on scaffold 48(Bfv1UA, Bfv2A: f_V2_138) and had 41 % identity to GLA of human prothrombin. Back-search indicated coagulation factor VII, prothrombin and proline-rich Gla protein 3 (PRRG3) in human and proteins described as coagulation factors VII and X in lamprey. The lancelet GLA domains were 41 and 31 % similar to lone GLA of acorn worm, respectively. Both of them were accompanied by EGF domains at the C-terminals. Any protease and kringle domains were present in neither acorn worm nor amphioxus K-dependent proteins.
Kringle Domains
Kringle domains were common in the sea urchin, acorn worm, and abundant in amphioxus, returning over 30, 60, and 100 hits, respectively, often repeating in the same gene. Acorn worm and sea urchin had many proteins described as “plasminogen-like”, “similar to plasminogen”, or “apolipoprotein(a)-like”. They showed 34–45 % similarity with human plasminogen kringles, some of them were proteases with zinc-dependent metalloprotease domains, but only seven proteins had kringles connected with serine protease domain at N-terminals.
Scaffold 396 Protease Prediction and Molecular Docking
The protein found in amphioxus on scaffold 396 was chosen to predict its hypothetical spatial structure, because of its close 42 % similarity to thrombin serine domain of human. The most imminent homologs in acorn worm and sea urchin were also modeled together with thrombin of Japanese lamprey (Lethenteron japonicum) and zebra fish (Danio reiro) as well as three hypothetical predecessors (vertebrate, chordate, and deuterostome ancestor) reconstructed by ANCESCON (Fig. 5). According to the predicted spatial structure, the chordate ancestor was much more similar to vertebrate ancestor and thrombin of vertebrates than to other predicted proteins. Common ancestor of tested deuterostomes was more closely related to sea urchin and acorn worm (Fig. 5).
Thrombin evolution. Hypothetical evolution of spatial structure of thrombin-like serine protease from hypothetical ancestors to descendants: lancelet, acorn worm, sea urchin and thrombin in vertebrates based on predicted structures and known human thrombin 1FPH structure. Software used ANCESCON, Autodock Tools v 1.5.6rc1
Docking of the conserved fragments of fibrinopeptide A, hirudin and a part of PAR-1 predicted by ANCESCON amphioxus protease returned 4, 6 and 4 conformations to 9, respectively, in areas similar to those which are present in the native human thrombin. However, the ligands were bound in distinct configurations (Fig. 6).
Binding of ligands. Comparison of in vitro binding of three physiological ligands with thrombin (top pair) and in silico docking with lancelet thrombin-like protease (bottom pair), without and with ligand (ball structures). The amino acid residues on the surface of binding place are shaded with squares and their sequence is written below. Ligands a fibrinopeptide A, b hirudin, c peptide NRS. Software used Autodock Vina 1.0, Autodock Tools v 1.5.6rc1
Discussion
The last common ancestor of all chordates probably lived before the Cambrian period, 550 million years ago. According to molecular phylogeny cephalochordates diverged first, before the split between urochordates (ascidians like sea squirt C. intestinalis) and vertebrates (Putnam et al. 2008). Although the sea squirt genome examination showed some slight signs of “precoagulation” factors (Jiang and Doolittle 2003), modern ascidians, even more related molecularly to vertebrates, albeit diverse morphologically, were simplified by gene loss, exon–intron loss and genome rearrangements (Putnam et al. 2008) and might lose many of “precoagulant” proteins. Gene content, exon–intron structure, and chromosomal organization make the amphioxus genome a better surrogate for the ancestral chordate genome (Putnam et al. 2008). Hemichordates like S. kowalevskii and echinoderms like S. purpuratus are the closest relatives of chordates and may serve as good reference point. Although our study confirms that there are no authentic coagulation factors in the amphioxus (Doolittle 2009) or in pre-chordates genomes, we found extremely high amount of different coagulation-like domains that shed some additional light on beginnings of hemostasis.
Thrombin-Like Serine Protease Domains in Non-Vertebrate Deuterostomes
Amphioxus genome is particularly rich in serine proteases domains. Some of them show distinct similarities to thrombin. The interesting case is a protein located on scaffold 396 (or XP_002587250 in NCBI nomination). It is 42 % identical to human thrombin, shows the high conservation in sequences characteristic for thrombin, lies close to thrombin on the phylogenetic tree, shares synteny with carnitine palmitoyltransferase, and it only slightly differs from vertebrate thrombin in hypothetical spatial structure (Figs. 2, 3, 4, 5, 6). The most striking feature of thrombin is its interaction with Na+. Na+ binding converts thrombin from the low activity slow (Na+-free) to the high activity fast (Na+-bound) form. At the physiological concentration of Na+ (140 mM) ratio of these two forms is 2:3. The fast form is responsible for the cleavage of fibrinogen, activation of coagulation factors V, VIII, XI, and XIII. The fast form is also responsible for the activation of PAR receptors leading to platelets activation and cell signaling. The slow form activates the anticoagulant protein C. The Na+ binding side is located between the 220-loop and the 186-loop and is within 5A from the side chain of Asp 189. (Di Cera 2003; Di Cera 2007; Di Cera 2008; Di Cera 2009; Di Cera 2011; Di Cera et al. 1995; Gandhi et al. 2011; Lane et al. 2005). The thrombin homolog found in amphioxus has conserved sequence in described region (Fig. 2). Amino acid Ser214 and Tyr225 are also conserved (except for the acorn worm where Y → F) as well as GDSGGP motif with catalytic triad Ser195 almost in the middle. The last mentioned residues have important role in the evolution of trypsin-like enzymes in the clotting, complement and developmental cascades (Krem and Di Cera 2002; Loof et al. 2011). The residue 225 is tyrosine or proline in 95 % of serine proteases from chymotrypsin subfamily. The tyrosine amino acid residue at this position is modern comparing to more ancestral proline residue. Tyrosine residue enables acquisition of Na+-binding and enhancement of activity, by controlling the conformation of surrounding reaction pocket. This new feature corresponds to secretion of such proteases into the extracellular fluid, were concentration of Na+ is higher than inside the cell, and to the evolutionary appearance of hemolymph with coagulation and complement function in more complex animals (Smith et al. 2001). The other important thing is conservation of exosite II, which is known to be responsible for platelet glycoprotein binding (Li et al. 2001). On the other hand, sequences corresponding to thrombin exosite I seem to be modified by the evolution much more. This protein has several close homologs not only in lancelet genome, but also in genomes of hemichordates and echinoderms. These features allow reconstructing the “pre-thrombin” evolution and building a tree-based on predicted spatial structures of thrombin homologs arranged together according to the known divergence of deuterostomes. It reveals that thrombin-like protein ancestor could be present already in chordate predecessor, afterwards evolved divergently in vertebrates and amphioxus (Fig. 5), but was lost in sea squirts as there is no such close homolog. The closest homolog C. intestinalis is only 36 % identical (more similar to prostasin-like, XP_002131103) and tunicates are currently considered to be closely related to vertebrates rather than to cephalochordates (Putnam et al. 2008). The mentioned hypothetical chordate protease could in turn evolve from protein that existed even before divergence of echinoderms (Fig. 5). Recent duplications in cephalochordates formed additional paralogs of the hypothetical predecessor, as could by demonstrated by proteins 75 and 70 % identical with the scaffold 396 protein (or XP_002587250) which are present in amphioxus genome. In evolutionary time scale 70–75 % similarity is a sizable relationship because such proteins like plasma prekallikrein and coagulation XI in human share 68 % identity and diverged probably after split of monotremes and viviparous mammals (Ponczek et al. 2008).
On the basis of our results, it can be concluded that the ability to cut fibrinogen had to occur in the descendants of described protein ancestor later as, despite some signs of fibrinogen-like genes (Doolittle 2012), there is no fibrinogen-like protein in non-vertebrate chordates which might have cleaved peptides to expose complementary knobs needed for polymerization.
Plasminogen-Like Serine Protease Domains and Lack of Functional Fibrin Polymerization
Another group of homologs are “plasminogen-like proteins” that can be found in the lancelet genome by bioinformatic search. One of them cloned from B. belcheri tsingtauense by Liu and Zhang (Liu and Zhang 2009) (92 % ortholog of XP_002596007 in B. floridae) was even called plasminogen (ACE86411). Could it be functional plasminogen as no true fibrinogen genes were found and no PAN domains in kringle proteases were present in amphioxus (Doolittle 2009; Doolittle 2011)? There are many fibrinogen-related proteins in amphioxus, as for instance multivalent pattern recognition receptor with a bacteriolytic activity cloned in B. belcheri (Fan et al. 2008) but none is true ortholog of vertebrate fibrinogen. A short in silico report has recently been published about probable fibrinogen-like proteins in C. intestinalis (Doolittle 2012). According to the Doolittle’s study sea squirt has three genes localized one behind the other resembling vertebrate fibrinogen chains. The proteins seems to have one RGD sequence in each of three chains, and binding sites in C-terminal domains (a feature characteristic for FRED domains), which could give a possibility to bind with cells that could aggregate at sites of injury. No N-terminal knobs and extensions with a possible site for cleavage by thrombin were found. A FReD domain of hypothetical product of one of this genes is actually quite similar to the vertebrate fibrinogen gamma FReD domain—48 % to bony fish—Tetraodon nigroviridis and 45 % to human, but three genes lie on the same strand of DNA (the genes can be found on GenBank: AABS01000073.1 or GenBank: EAAA01001286.1, which are complementary strands to each other.), not like genes of vertebrate fibrinogen, where Bbeta gene is located on opposite strand than Aalpha and gamma. The two other genes do not seem to be such similar giving less than 40 % resemblance within the FReD domain. It cannot be excluded these genes may signify some signs of multiplication of fibrinogen ancestral genes before splitting of sea squirt and vertebrates predecessors, but after divergence of lancelet, the more so that it has been found by Doolittle that the gene duplications leading to the evolution of the three Ciona polypeptides and the Bbeta and gamma chains of vertebrate fibrinogen occurred within the same general time frame.
It is known that sea squirt has several plasminogen-like proteases with attached kringle domains (Jiang and Doolittle 2003). No one can at the moment say wheather some of them could split the sea squirt cells clumped by hypothetical fibrinogen-like protein mediation. It seems that amphioxus also has fibrinogen-like proteins (Doolittle 2011). Some of them could have similar properties to the protein found in sea squirt. None of them, however, has fibrinopeptides and knobs recognized and cut thrombin (Doolittle 2012). This fact is consistent with week conservation of Exosite I of thrombin-like protein. The other interesting thing in all studied animals is the lack of close homologs of characteristic for thrombin PAR receptors. NCBI Blast returns only 27–28 % similar results that in back searches are close in 41–42 % to human somatostatin receptors. This in turn is in agreement with poor conservation of PAR binding sequences. On the other hand the conservation of exosite II, with its role in glycoprotein binding, may indicate the importance of the ancestor of thrombin in intercellular clumping (Di Cera 2008; Ponczek 2010a; Ponczek 2010b). Additionally, this hypothesis can be confirmed by conservation of RGD sequence in acorn worm in this region (Fig. 2).
Referring back to plasminogen, mentioned earlier facts, particularly 44 % similarity of protease domain to human plasminogen protease domain and the observation that ACE86411 in B. belcheri as well as XP_002596007 in B. floridae have tPA activation place (estimated in silico) and that the cloned protein is activated by uPA in vitro (Liu and Zhang 2009) suggest that lancelet “plasminogen” may be a descendant of a sole protein predecessor which could have existed in chordate common ancestor and could have given birth to functional plasminogen with multiplication of kringle domains after divergence of vertebrate ancestors.
Plasminogen Activators-Like Serine Protease Domains
Moreover, some additional proteases with two kringle domains or with one kringle domain in amphioxus genome were found. Nevertheless, none of them could have been a close relative to vertebrate tPA or uPA, because these homologs were more similar to serine proteases not belonging to hemostatic system. Potential related protease domain could be XP_002605710, which is 40 % identical with both human plasminogen activators. This amphioxus protein does not have kringle domains and it has nearest no kringle homologs in acorn worm and sea urchin. Taking into account that human tPA and uPA share 45 % similarity and are located on separate chromosomes (8 and 10, respectively in human), their splitting must have happened after divergence of vertebrates as an act of global genome duplication or chromosomal duplication.
Multiplication of GLA Domains
The duplication events are well illustrated by GLA domain multiplication. Furthermore, the number of GLA in the acorn worm genome seems to be a half of the number in the lancelet genome and the number of GLA in the lancelet genome is a half of the number of corresponding domains in the sea squirt genome. This subtly outlines how domains needed for the formation of functional factors of hemostasis could be multiplied by succeeding duplications.
The Emergence of Hemostasis Proteases
Although no authentic coagulation factors could be found in any of non-vertebrate chordates and pre-chordate animals, the comparison of genomes of these organisms with vertebrates shed light on the possible scenario of assembling of proteases in hemostasis evolution. Our results partly contradict the putative model of evolution of proteases involved in generation and destruction of fibrin clots presented by Jiang and Doolittle (Jiang and Doolittle 2003) and also described later (Doolittle 2009). The authors suggested the existence of one ancestral four- or three-kringle root protease that gave birth to prothrombin, plasminogen, both plasminogen activators and other coagulation proteases. Our results do not confirm existence of one ancestral protease for mentioned hemostatic factors that could possess kringle domains but rather indicate the presence of at least three different proteases in primeval deuterostomes. The main arguments against hypothesis of one 3 or 4 kringle predecessor are too large differences in similarities of protease domains between different factors and too much similarity with the same type occurring in vertebrates (Table 2) as well as arrangement or lack of additional domains in tested organisms. We hypothesize that in common cephalochordate and vertebrate ancestor minimum three serine proteases could be present that gave birth to IG-like, no kringle and two kringle proteases in lancelets, and accordingly prothrombin, plasminogen activators, and plasminogen/HFG in vertebrates (Fig. 7). No other protease hemostasis factors had such clear equivalents. It is possible that in common predecessor of vertebrates the ancestor proteases could diverge into present coagulation and fibrinolysis factors after merging with proper N-terminal domains and some adjuvant duplications leading to appearing of cofactors and fibrinogen. The predecessor of thrombin could exists even 710–780 million years ago (Fig. 3), which is the time close to divergence of protostomes and deuterostomes (Peterson et al. 2008). Our findings are consistent with hypothesis of Kulman et al. (Kulman et al. 2006). They suggest Gla-EGF2-serine protease prototype arose independently of prothrombin. This could probably mean that the fourth root serine protease could have existed that gave Gla-EGF2-serine protease coagulation factors (Table 4).
Diagrammatic depiction of the deuterostome evolutionary phylogenetic relationships and proposed history of hemostatic-like protease evolution in cephalochordates and vertebrates. a Deuterostome evolutionary tree with proposed history of evolutionary changes (based on McClay 2011). ECH—Echinodermata (e.g., sea urchin), HEM—Hemidermata (e.g., acorn worm), CEP—Cephalochordata (e.g., amphioxus), URO—Urochordata (e.g., sea squirt), CHO—Chordata (e.g., vertebrates). Duplications that led to three putative serine proteases—*, shuffling of domains and further duplications that led to amphioxus proteases, acquisition of new domains and duplications which led to the formation of vertebrate hemostatic factors—***. b Putative divergence of serine proteases of hemostasis and their homologs (known domain arrangement based on sequence PROSITE anticipation—http://www.expasy.ch/prosite). The common chordate ancestor could have had at least three proteins that could have been predecessors of main vertebrate proteases of hemostasis: the first for prothrombin and possibly other extrinsic pathway proteases (1), the second for plasminogen activators (2) and the third for plasminogen (3)
It has been recently known that some serine proteases occurring in urochordates like Botryllus schlosseri are homologous to vertebrate blood coagulation proteases and participate in reactions provoked by meeting of different colonies what leads to cell clumping at the sites of contact (Oren et al. 2008). This may indicate a primarily defensive role, originally fulfilled by the cascade activated serine proteases, which subsequently evolved in vertebrates into the complement system and hemostasis. The future research in the field of molecular biology would localize the place of expression and explain the role of described proteases in pre-chordate organism’s further developing understanding of the evolution of hemostasis.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94
Cai W, Pei J, Grishin NV (2004) Reconstruction of ancestral protein sequences and its applications. BMC Evol Biol 4:33
Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J (2005) ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423
Cera Di, Guinto ER, Vindigni A, Dang QD, Ayala YM, Wuyi M, Tulinsky A (1995) The Na+ binding site of thrombin. J Biol Chem 270:22089–22092
Davidson CJ, Hirt RP, Lal K, Snell P, Elgar G, Tuddenham EG, McVey JH (2003a) Molecular evolution of the vertebrate blood coagulation network. Thromb Haemost 89:420–428
Davidson CJ, Tuddenham EG, McVey JH (2003b) 450 million years of hemostasis. J Thromb Haemost 1:1487–1494
De Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:362–365
Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De TA, Davidson B, Di GA, Gelpke M, Goodstein DM et al (2002) The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298:2157–2167
Di Cera E (2003) Thrombin interactions. Chest 124:11–17
Di Cera E (2007) Thrombin as procoagulant and anticoagulant. J Thromb Haemost 5(suppl 1):196–202
Di Cera E (2008) Thrombin. Mol Aspects Med 29:203–254
Di Cera E (2009) Serine proteases. IUBMB Life 61:115–510
Di Cera E (2011) Thrombin as an anticoagulant. Prog Mol Biol Transl Sci 99:145–184
Doolittle RF (1993) The evolution of vertebrate blood coagulation: a case of Yin and Yang. Thromb Haemost 70:24–28
Doolittle RF (2009) Step-by-step evolution of vertebrate blood coagulation. Cold Spring Harb Symp Quant Biol 74:35–40
Doolittle RF (2011) Coagulation in vertebrates with a focus on evolution and inflammation. J Innate Immun 3:9–16
Doolittle RF (2012) The protochordate Ciona intestinalis has a protein like full-length vertebrate fibrinogen. J Innate Immun 4:219–222
Doolittle RF, Feng D-F (1987) Reconstructing the evolution of vertebrate blood coagulation from a consideration of the amino acid sequences of clotting proteins. Cold Spring Harbor Symp Quant Biol 52:869–874
Doolittle RF, Jiang Y, Nand J (2008) Genomic evidence for a simpler clotting scheme in jawless vertebrates. J Mol Evol 66:185–196
Escriva H, Manzon L, Youson J, Laudet V (2002) Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. Mol Biol Evol 19:1440–1450
Fan C, Zhang S, Chao LLY (2008) Fibrinogen-related protein from amphioxus Branchiostoma belcheri is a multivalent pattern recognition receptor with a bacteriolytic activity. Mol Immunol 45:3338–3346
Felsenstein J (1989) PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5:164–166
Gandhi PS, Chen Z, Di Cera E (2010) Crystal structure of thrombin bound to the uncleaved extracellular fragment of PAR1. J Biol Chem 285:15393–15398
Gandhi PS, Chen Z, Appelbaum E, Zapata F, Di Cera E (2011) Structural basis of thrombin-protease-receptor interactions. IUBMB Life 63:375–382
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89:10915–10919
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Jiang Y, Doolittle RF (2003) The evolution of vertebrate blood coagulation as viewed from a comparison of puffer fish and sea squirt genomes. Proc Natl Acad Sci USA 100:7527–7532
Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Res 37:D387–D392
Kimura A, Ikeo K, Nonaka M (2009) Evolutionary origin of the vertebrate blood complement and coagulation systems inferred from liver EST analysis of lamprey. Dev Comp Immunol 33:77–87
Krem MM, Di Cera E (2002) Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem Sci 27:67–74
Kulman JD, Harris JE, Nakazawa N, Ogasawara M, Satake M, Davie EW (2006) Vitamin K dependent proteins in Ciona intestinales, a basal chordate lacking a blood coagulation cascade. Proc Natl Acad Sci USA 03:15794–15799
Lane DA, Philippou H, Huntington JA (2005) Directing thrombin. Blood 106:2605–2612
Letunic I, Bork P (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128
Li CQ, Vindigni A, Sadler JE, Wardell MR (2001) Platelet glycoprotein Ib alpha binds to thrombin anion-binding exosite II inducing allosteric changes in the activity of thrombin. J Biol Chem 276:6161–6168
Liu M, Zhang S (2009) A kringle-containing protease with plasminogen-like activity in the basal chordate Branchiostoma belcheri. Biosci Rep 29:385–395
Loof TG, Schmidt O, Herwald H, Theopold U (2011) Coagulation systems of invertebrates and vertebrates and their roles in innate immunity: the same side of two coins? J Innate Immun 3:34–40
Mathews II, Padmanabhan KP, Ganesh V, Tulinsky A, Ishii M, Chen J, Turck CW, Coughlin SR, Fenton JW (1994) Crystallographic structures of thrombin complexed with thrombin receptor peptides: existence of expected and novel binding modes. Biochemistry 33:3266–3279
McClay DR (2011) Evolutionary crossroads in developmental biology: sea urchins. Development 138:2639–2648
Nonaka M, Yoshizaki F (2004) Evolution of the complement system. Mol Immunol 40:897–902
Oren M, Escande MI, Paz G, Fishelson Z, Rinkevich B (2008) Urochordate histoincompatible interactions activate vertebrate-like coagulation system components. PLoS ONE 3:e3123
Peterson KJ, Cotton JA, Gehling JG, Pisani D (2008) The Ediacaran emergence of bilaterians: congruence between the genetic and the geological fossil records. Philos Trans R Soc Lond B Biol Sci 63:1435–1443
Ponczek MB (2010a) Evolution of vertebrate hemostatic system. Kosmos 59:83–90
Ponczek MB (2010b) The contact factors of hemostasis: examination of molecular evolution and new therapeutic perspectives. Postepy Biochem 56:67–74
Ponczek MB, Gailani D, Doolittle RF (2008) Evolution of the contact phase of vertebrate blood coagulation. J Thromb Haemost 6:1876–1883
Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK et al (2008) The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071
Riddel JP Jr, Aouizerat BE, Miaskowski C, Lillicrap DP (2007) Theories of blood coagulation. J Pediatr Oncol Nurs 24:123–131
Schenone M, Furie BC, Furie B (2004) The blood coagulation cascade. Curr Opin Hematol 11:272–277
Smith LC, Clow LA, Terwilliger DP (2001) The ancestral complement system in sea urchins. Immunol Rev 180:16–34
Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD et al (2006) The genome of the sea urchin Strongylocentrotus purpuratus. Science 314:941–952
Sotriffer CA, Flader W, Winger RH, Rode BM, Liedl KR, Varga JM (2000) Automated docking of ligands to antibodies: methods and applications. Methods 20:280–291
Stubbs MT, Oschkinat H, Mayr I, Huber R, Angliker H, Stone SR, Bode W (1992) The interaction of thrombin with fibrinogen. A structural basis for its specificity. Eur J Biochem 206:187–195
Suzuki MM, Satoh N, Nonaka M (2002) C6-lile and C3-like molecules from the cephalochordate, amphioxus, suggest a cytolytic complement system in invertebrates. J Mol Evol 54:671–679
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Acknowledgments
We are grateful to Professor Russell Doolittle and Professor Barbara Wachowicz for scientific care and advice as well as to the European Molecular Biology Organization for founding the Short Term Fellowship ASTF 83-2008 for Michal Ponczek and also to the University of Lodz for grant No. 506/810. Without all of these the work would not probably be created.
Conflict of interest
The authors declare no competing financial interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
239_2012_9509_MOESM1_ESM.doc
Appendix 1. Sequences (Fig. 2), sequences and results of phylogenetic analysis (Fig. 3). Supplementary material 1 (DOC 243 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ponczek, M.B., Bijak, M.Z. & Nowak, P.Z. Evolution of Thrombin and Other Hemostatic Proteases by Survey of Protochordate, Hemichordate, and Echinoderm Genomes. J Mol Evol 74, 319–331 (2012). https://doi.org/10.1007/s00239-012-9509-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-012-9509-0