Abstract
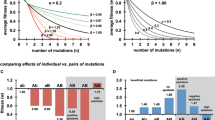
In previous work (Betancourt, Genetics 181:1535, 2009), I propagated three large laboratory populations of an RNA phage (MS2) as they adapted to a controlled laboratory environment. These populations were large enough so that evolution might be expected to be mostly repeatable, but they nevertheless fixed different suites of mutations over the course of the experiment. Here, I investigate one possible explanation for these results: epistasis, in which the effect of a mutation depends on its genetic background, may have prevented populations with different initial substitutions from fixing the same set of subsequent mutations. I show that two mutations that previously occurred in different genetic backgrounds are beneficial on either background. This result suggests that sign epistasis—in which a mutation is beneficial on one background, but deleterious on another—is not the cause of different evolutionary trajectories observed in the Betancourt (2009) experiment. However, they can be explained by either magnitude epistasis—in which mutations have stronger or weaker beneficial effects depending on the background—or by the simultaneous fixation of multiple beneficial mutations. Surprisingly, the large populations of the previous experiment showed less parallel evolution than the small populations of this experiment, which lends support to the fixation of multiple beneficial mutations contributing to the patterns seen in both experiments.

Similar content being viewed by others
References
Belshaw R, Gardner A, Rambaut A, Pybus OG (2008) Pacing a small cage: mutation and RNA viruses. Trends Ecol Evol 23:188–193
Betancourt AJ (2009) Genomewide patterns of substitution in adaptively evolving populations of the RNA bacteriophage MS2. Genetics 181:1535
Bollback JP, Huelsenbeck JP (2007) Clonal interference is alleviated by high mutation rates in large populations. Mol Biol Evol 24:1367–1406
Bollback JP, Huelsenbeck JP (2009) Parallel genetic evolution within and between bacteriophage species with varying degrees of divergence. Genetics 181:225–234
Bull JJ, Badgett MR, Wichman HA (2000) Big-benefit mutations in a bacteriophage inhibited with heat. Mol Biol Evol 17:942–950
Burch C, Chao L (2000) Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature 406:625–628
Coyne JA, Orr HA (2004) Speciation. Sinauer Associates, Sunderland, MA
Coyne JA, Barton NH, Turelli M (2000) Is Wright’s shifting balance process important in evolution? Evol Int J Org Evol 54:306–317
Crill WD, Wichman HA, Bull JJ (2000) Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154:27–37
Cuevas JM, Elena SF, Moya A (2002) Molecular basis of adaptive convergence in experimental populations of RNA viruses. Genetics 162:533–542
Desai MM, Fisher DS, Murray AW (2007) The speed of evolution and maintenance of variation in asexual populations. Curr Biol 17:385–394
Drake JW (1993) Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci USA 90:4171–4175
Drake JW, Charlesworth B, Charlesworth D, Crow JF (1998) Rates of spontaneous mutation. Genetics 148:1667–1686
Gerrish PJ, Lenski RE (1998) The fate of competing beneficial mutations in an asexual population. Genetica 102–103:127–144
Goodnight CJ, Wade MJ (2000) The ongoing synthesis: a reply to Coyne, Barton, and Turelli. Evol Int J Org Evol 54:317–324
Holder KK, Bull JJ (2001) Profiles of adaptation in two similar viruses. Genetics 159:1393–1404
Holmes EC (2003) Error thresholds and the constraints to RNA virus evolution. Trends Microbiol 11:543–546
Johnson T, Barton NH (2005) The effect of deleterious alleles on adaptation in asexual populations. Genetics 162:395–411
Kern AD, Kondrashov FA (2004) Mechanisms and convergence of compensatory evolution in mammalian mitochondrial tRNAs. Nat Genet 36:1207–1212
Kim Y, Orr HA (2005) Adaptation in sexuals versus asexuals: clonal interference and the Fisher-Muller model. Genetics 171:1377–1386
Klovins J, Tsareva NA, de Smit MH, Berzins V, van Duin J (1997) Rapid evolution of translational control mechanisms in RNA genomes. J Mol Biol 265:372–384
Kondrashov AS, Sunyaev S, Kondrashov FA (2002) Dobzhansky-Muller incompatibilities in protein evolution. Proc Natl Acad Sci USA 99:14878–14883
Kulathinal RJ, Bettencourt BR, Hartl DL (2004) Compensated deleterious mutations in insect genomes. Science 306:1553–1554
Mo H, Stamatatos L, Ip JE, Barbas CF, Parren PW, Burton DR, Moore JP, Ho DD (1997) Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. off. J Virol 71:6869–6874
Peck JR (1994) A ruby in the rubbish: beneficial mutations, deleterious mutations and the evolution of sex. Genetics 137:597–606
Poon A, Chao L (2005) The rate of compensatory mutation in the DNA bacteriophage phiX174. Genetics 170:989–999
Poon A, Davis BH, Chao L (2005) The coupon collector and the suppressor mutation: estimating the number of compensatory mutations by maximum likelihood. Genetics 170:1323–1332
Rokyta DR, Joyce P, Caudle SB, Wichman HA (2005) An empirical test of the mutational landscape model of adaptation using a single-stranded DNA virus. Nat Genet 37:441–444
Sanjuan R, Moya A, Elena SF (2004) The contribution of epistasis to the architecture of fitness in an RNA virus. Proc Natl Acad Sci USA 101:15376–15379
The Chimp Genome Sequencing Consortium (2005) Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87
Unckless RL, Orr HA (2009) Dobzhansky-Muller incompatibilities and adaptation to a shared environment. Heredity 102:214–217
Wahl LM, Krakauer DC (2000) Models of experimental evolution: the role of genetic chance and selective necessity. Genetics 156:1437–1448
Weinreich DM, Watson RA, Chao L (2005) Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evol Int J Org Evol 59:1165–1174
Weinreich DM, Delaney NF, Depristo MA, Hartl DL (2006) Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312:111–114
Welch JW, Waxman D (2005) The nk model and population genetics. J Theor Biol 234:329–340
Wichman HA, Badgett MR, Scott LA, Boulianne CM (1999) Different trajectories of parallel evolution during viral adaptation. Science 285:422–424
Wood TE, Burke JM, Rieseberg LH (2005) Parallel genotypic evolution: when evolution repeats itself. Genetica 123:157–170
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Acknowledgments
Thanks to J. Bollback, S. Collins, K. Dyer, H. Kuehne, D. Presgraves, and J. Welch for helpful discussion and to L. Yanoso for technical help. I particularly thank A. Orr and J. Huelsenbeck for helpful discussion and encouragement during the course of this experiment. This work was funded in part by a National Science Foundation (NSF) Doctoral Dissertation Improvement grant to A.J.B. and H. A. Orr.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Betancourt, A.J. Lack of Evidence for Sign Epistasis Between Beneficial Mutations in an RNA Bacteriophage. J Mol Evol 71, 437–443 (2010). https://doi.org/10.1007/s00239-010-9397-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-010-9397-0