Abstract
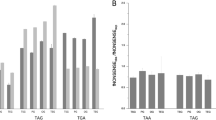
Understanding the cause of the changes in the amino acid composition of proteins is essential for understanding the evolution of protein functions. Since the early 1970s, it has been known that the frequency of some amino acids in protein sequences is increasing and that of others is decreasing. Recently, it was found that the trends of amino acid changes were similar in 15 taxa representing Bacteria, Archaea, and Eukaryota. However, the cause of this similarity in the trend of the gains and losses of amino acids continued to be debated. Here, we show that this trend of the gain and loss of amino acids can be simply explained by CpG hypermutability. We found that the frequency of amino acids coded by codons with TpG dinucleotides and those with CpA dinucleotides is increasing, while that of amino acids coded by codons with CpG dinucleotides is decreasing. We also found that organisms that lack DNA methyltransferase show different trends of the gain and loss of amino acids. DNA methyltransferase methylates CpG dinucleotides and induces CpG hypermutability. The incorporation of CpG hypermutability into models of protein evolution will improve studies on protein evolution in different organisms.



Similar content being viewed by others
References
Bird AP (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8:1499–1504
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Goldstein RA, Pollock DD (2006) Observations of amino acid gain and loss during protein evolution are explained by statistical bias. Mol Biol Evol 23:1444–1449
Hobolth A, Nielsen R, Wang Y, Wu F, Tanksley SD (2006) CpG + CpNpG analysis of protein-coding sequences from tomato. Mol Biol Evol 23:1318–1323
Hurst LD, Fell EJ, Rocha EPC (2006) Protein evolution: causes of trends in amino-acid gain and loss. Nature 442:E11–E12
Huttley GA (2004) Modeling the impact of DNA methylation on the evolution of BRCA1 in mammals. Mol Biol Evol 21:1760–1768
Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S (2005) A universal trend of amino acid gain and loss in protein evolution. Nature 433:633–638
Jukes TH (1978) Codons and nearest-neighbor nucleotide pairs in mammalian messenger RNA. J Mol Evol 11:121–127
Kimura M (1968) Evolutionary rate at the molecular level. Nature 217:624–626
Kosiol C, Holmes I, Goldman N (2007) An empirical codon model for protein sequence evolution. Mol Biol Evol 24:1464–1479
Lunter G, Hein J (2004) A nucleotide substitution model with nearest-neighbour interactions. Bioinformatics 20(Suppl 1):I216–I223
McDonald JH (2006) Apparent trends of amino acid gain and loss in protein evolution due to nearly neutral variation. Mol Biol Evol 23:240–244
Ohta T (1992) The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23:263–286
Ponger L, Li WH (2005) Evolutionary diversification of DNA methyltransferases in eukaryotic genomes. Mol Biol Evol 22:1119–1128
R Development Core Team (2007) R: a language and environment for statistical computing. R Development Core Team, Vienna, Austria
Riggins GJ, Lokey LK, Chastain JL, Leiner HA, Sherman SL, Wilkinson KD, Warren ST (1992) Human genes containing polymorphic trinucleotide repeats. Nat Genet 2:186–191
Scarano E, Iaccarino M, Grippo P, Parisi E (1967) The heterogeneity of thymine methyl group origin in DNA pyrimidine isostichs of developing sea urchin embryos. Proc Natl Acad Sci USA 57:1394–1400
Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H (2000) Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81–86
Tweedie S, Charlton J, Clark V, Bird A (1997) Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol 17:1469–1475
Wang Y, Rocha EP, Leung FC, Danchin A (2004) Cytosine methylation is not the major factor inducing CpG dinucleotide deficiency in bacterial genomes. J Mol Evol 58:692–700
Wootton JC (1994) Sequences with ‘unusual’ amino acid compositions. Curr Opin Struct Biol 4:413–421
Zuckerkandl E, Derancourt J, Vogel H (1971) Mutational trends and random processes in the evolution of informational macromolecules. J Mol Biol 59:473–490
Acknowledgments
This project was supported by grants from the Kazusa DNA Research Institute. This study was also supported by the Collaboration of Regional Entities for the Advancement of Technological Excellence (CREATE) Program of the Japan Science and Technology (JST) Corporation as well as by the National Project on “Next-Generation Integrated Living Matter Simulation” of the Ministry of Education, Culture, Sports, Science and Technology (MEXT). Computation time was provided by the Super Computer System, Human Genome Center, Institute of Medical Science, University of Tokyo.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Misawa, K., Kamatani, N. & Kikuno, R.F. The Universal Trend of Amino Acid Gain–Loss is Caused by CpG Hypermutability. J Mol Evol 67, 334–342 (2008). https://doi.org/10.1007/s00239-008-9141-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-008-9141-1