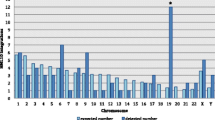
Abstract
We and others recently identified an almost-intact human endogenous retrovirus (HERV), termed HERV-K(HML-2.HOM), that is usually organized as a tandem provirus. Studies on HERV proviral loci commonly rely on the analysis of single alleles being taken as representative for a locus. We investigated the frequency of HERV-K(HML-2.HOM) single and tandem alleles in various human populations. Our analysis revealed that another HERV-K(HML-2) locus, the so-called HERV-K(II) provirus, is also present as a tandem provirus allele in the human population. Proviral tandem formations were identified in various nonhuman primate species. We furthermore examined single nucleotide polymorphisms (SNPs) within the HERV-K(HML-2.HOM) proviral gag, prt, and pol genes, which all result in nonsense mutations. We identified four proviral haplotypes displaying different combinations of gag, prt, and pol SNPs. Haplotypes harboring completely intact proviral genes were not found. For the left provirus of the tandem arrangement a haplotype displaying intact gag and prt genes and a mutated pol was found in about two-thirds of individuals from different ethnogeographic origins. The same haplotype was always found in the right provirus. The various haplotypes point toward multiple recombination events between HERV-K(HML-2.HOM) proviruses. Based on these findings we derive a model for the evolution of the proviral locus since germ line integration.






Similar content being viewed by others
References
Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J (1999) Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol 9:861–868
de Parseval N, Heidmann T (1998) Physiological knockout of the envelope gene of the single-copy ERV-3 human endogenous retrovirus in a fraction of the Caucasian population. J Virol 72:3442–445
Hughes JF, Coffin JM (2001) Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat Genet 29:487–489
Hughes JF, Coffin JM (2004) Human endogenous retrovirus K solo–LTR formation and insertional polymorphisms: Implications for human and viral evolution. Proc Natl Acad Sci USA 101:1668–1672
International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Kambhu S, Falldorf P, Lee JS (1990) Endogenous retroviral long terminal repeats within the HLA-DQ locus. Proc Natl Acad Sci USA 87:4927–4931
Kinjo Y, Matsuura N, Yokota Y, Ohtsu S, Nomoto K, Komiya I, Sugimoto J, Jinno Y, Takasu N (2001) Identification of nonsynonymous polymorphisms in the superantigen-coding region of IDDMK1,2 22 and a pilot study on the association between IDDMK1,2 22 and type 1 diabetes. J Hum Genet 46:712–716
Li WH, Gu Z, Wang H, Nekrutenko A (2001) Evolutionary analyses of the human genome. Nature 409:847–849
Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J (1995) Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J Virol 69:141–149
Lower R, Lower J, Kurth R (1996) The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA 93:5177–5184
Macfarlane C, Simmonds P (2004) Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J Mol Evol 59:642–656
Mayer J, Meese E, Mueller-Lantzsch N (1997a) Chromosomal assignment of human endogenous retrovirus K (HERV-K) env open reading frames. Cytogenet Cell Genet 79:157–161
Mayer J, Meese E, Mueller-Lantzsch N (1997b) Multiple human endogenous retrovirus (HERV-K) loci with gag open reading frames in the human genome. Cytogenet Cell Genet 78:1–5
Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E (1999) An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet 21:257–258
Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF (1989) Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516
Ono M, Yasunaga T, Miyata T, Ushikubo H (1986) Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol 60:589–598
Reus K, Mayer J, Sauter M, Scherer D, Muller-Lantzsch N, Meese E (2001) Genomic organization of the human endogenous retrovirus HERV-K(HML-2HOM) (ERVK6) on chromosome 7. Genomics 72:314–20
Stoye JP (2001) Endogenous retroviruses: still active after all these years? Curr Biol 11:R914–R916
Sugimoto J, Matsuura N, Kinjo Y, Takasu N, Oda T, Jinno Y (2001) Transcriptionally active HERV-K genes: identification, isolation, and chromosomal mapping. Genomics 72:137–144
Tonjes RR, Czauderna F, Kurth R (1999) Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J Virol 73:9187–95
Tonjes RR, Lower R, Boller K, Denner J, Hasenmaier B, Kirsch H, Konig H, Korbmacher C, Limbach C, Lugert R, Phelps RC, Scherer J, Thelen K, Lower J, Kurth R (1996) HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol 13:S261–S267
Turner G, Barbulescu M, Su M, Jensen–Seaman MI, Kidd KK, Lenz J (2001) Insertional polymorphisms of full–length endogenous retroviruses in humans. Curr Biol 11:1531–1535
Acknowledgments
This work was supported by DFG Research Grants Ma2298/2-1 and Me917/16-1 to J.M. and E.M., respectively. Genomic DNA from Southeast Asian individuals was kindly provided by John Nicholls (University of Hongkong). Hans Zischler (University of Mainz, Germany) and Werner Schempp (University of Freiburg, Germany) generously provided primate genomic DNAs. We are grateful to Jochem König (University of Saarland, Germany) for help with statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor : Dr. Martin Kreitman]
Rights and permissions
About this article
Cite this article
Mayer, J., Stuhr, T., Reus, K. et al. Haplotype Analysis of the Human Endogenous Retrovirus Locus HERV-K(HML-2.HOM) and Its Evolutionary Implications. J Mol Evol 61, 706–715 (2005). https://doi.org/10.1007/s00239-005-0066-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0066-7