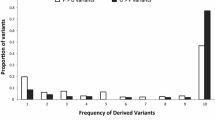
Abstract
One of the principal goals of population genetics is to understand the processes by which genetic variation within species (polymorphism) becomes converted into genetic differences between species (divergence). In this transformation, selective neutrality, near neutrality, and positive selection may each play a role, differing from one gene to the next. Synonymous nucleotide sites are often used as a uniform standard of comparison across genes on the grounds that synonymous sites are subject to relatively weak selective constraints and so may, to a first approximation, be regarded as neutral. Synonymous sites are also interdigitated with nonsynonymous sites and so are affected equally by genomic context and demographic factors. Hence a comparison of levels of polymorphism and divergence between synonymous sites and amino acid replacement sites in a gene is potentially informative about the magnitude of selective forces associated with amino acid replacements. We have analyzed 56 genes in which polymorphism data from D. simulans are compared with divergence from a reference strain of D. melanogaster. The framework of the analysis is Bayesian and assumes that the distribution of selective effects (Malthusian fitnesses) is Gaussian with a mean that differs for each gene. In such a model, the average scaled selection intensity (γ =N e s) of amino acid replacements eligible to become polymorphic or fixed is −7.31, and the standard deviation of selective effects within each locus is 6.79 (assuming homoscedasticity across loci). For newly arising mutations of this type that occur in autosomal or X-linked genes, the average proportion of beneficial mutations is 19.7%. Among the amino acid polymorphisms in the sample, the expected average proportion of beneficial mutations is 47.7%, and among amino acid replacements that become fixed the average proportion of beneficial mutations is 94.3%. The average scaled selection intensity of fixed mutations is +5.1. The presence of positive selection is pervasive with the single exception of kl-5, a Y-linked fertility gene. We find no evidence that a significant fraction of fixed amino acid replacements is neutral or nearly neutral or that positive selection drives amino acid replacements at only a subset of the loci. These results are model dependent and we discuss possible modifications of the model that might allow more neutral and nearly neutral amino acid replacements to be fixed.
Similar content being viewed by others
References
MD Adams SE Celniker RA Holt et al. (2000) ArticleTitleThe genome sequence of Drosophila melanogaster. Science 287 2185–2195 Occurrence Handle10.1126/science.287.5461.2185 Occurrence Handle10731132
H Akashi (1995) ArticleTitleInferring weak selection from patterns of polymorphism and divergence at “silent” sites in Drosophila DNA. Genetics 139 1067–1076
CD Bustamante R Nielsen DL Hartl (2002a) ArticleTitleA maximum likelihood method for analyzing pseudogene evolution: Implications for silent site evolution in humans and rodents. Mol Biol Evol 19 110–117 Occurrence Handle1:CAS:528:DC%2BD38XhtFCiug%3D%3D
CD Bustamante R Nielsen SA Sawyer KM Olsen MD Purugganan DL Hartl (2002b) ArticleTitleThe cost of inbreeding in Arabidopsis. Nature 416 531–534 Occurrence Handle1:CAS:528:DC%2BD38XivVOntr0%3D
BP Carlin TA Louis (2000) Bayes and empirical Bayes methods for data analysis. Chapman & Hall London
B Charlesworth D Charlesworth (1997) ArticleTitleRapid fixation of deleterious alleles can be caused by Muller’s ratchet. Genet Res 70 63–73 Occurrence Handle10.1017/S0016672397002899 Occurrence Handle1:STN:280:DyaK1c%2FjsFKjtQ%3D%3D Occurrence Handle9369098
JM Comeron M Kreitman (2002) ArticleTitlePopulation, evolutionary and genomic consequences of interference selection. Genetics 161 389–410
WJ Ewens (1972) ArticleTitleThe sampling theory of selectively neutral alleles. Theor Popul Biol 3 87–112 Occurrence Handle4667078
JC Fay C-I Wu (2000) ArticleTitleHitchhiking under positive Darwinian selection. Genetics 155 1405–1413 Occurrence Handle1:STN:280:DC%2BD3cvgsVSntQ%3D%3D Occurrence Handle10880498
JC Fay GJ Wyckoff C-I Wu (2002) ArticleTitleTesting the neutral theory of molecular evolution with genomic data from Drosophila. Nature 415 1024–1026 Occurrence Handle10.1038/4151024a Occurrence Handle11875569
Y-X Fu W-H Li (1993) ArticleTitleStatistical tests of neutrality of mutations. Genetics 133 693–709 Occurrence Handle1:STN:280:ByyB3c3kvVU%3D Occurrence Handle8454210
N Galtier F Depaulis NH Barton (2000) ArticleTitleDetecting bottlenecks and selective sweeps from DNA sequence polymorphism. Genetics 155 981–987 Occurrence Handle1:CAS:528:DC%2BD3cXkslyqurc%3D Occurrence Handle10835415
A Gelman JS Carlin HS Stern DB Rubin (1997) Bayesian data analysis. Chapman & Hall London
R Gilks S Richardson DJ Spiegelhalter (1996) Markov chain Monte Carlo in practice. Chapman & Hall London
JH Gillespie (2000) ArticleTitleGenetic drift in an infinite population. The pseudohitchhiking model. Genetics 155 909–919 Occurrence Handle1:STN:280:BymB28rlsVI%3D Occurrence Handle8685218
SJ Gould (1989) ArticleTitleTires to sandals: As we strive to understand nature, do we seek truth or solace? Nat Hist 98 8–15
DL Hartl AG Clark (1997) Principles of population genetics. Sinauer Associates Sunderland, MA
DL Hartl CH Taubes (1996) ArticleTitleCompensatory nearly neutral mutations: Selection without adaptation. J Theor Biol 182 303–309 Occurrence Handle10.1006/jtbi.1996.0168 Occurrence Handle1:CAS:528:DyaK28XntValsrg%3D Occurrence Handle8944162
DL Hartl EN Moriyama SA Sawyer (1994) ArticleTitleSelection intensity for codon bias. Genetics 138 227–234 Occurrence Handle1:STN:280:ByqD1MrgsV0%3D Occurrence Handle8001789
RR Hudson (1990) Gene geneologies and the coalescent process. D Futuyma J Antonovics (Eds) Oxford surveys in evolutionary biology, Oxford University Press Oxford 1–44
RR Hudson K Bailey D Skarecky J Kwiatowski FJ Ayala (1994) ArticleTitleEvidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136 1329–1340 Occurrence Handle1:CAS:528:DyaK2cXmslOjtLc%3D Occurrence Handle8013910
Y Kim W Stephan (2000) ArticleTitleJoint effects of genetic hitchhiking and background selection on neutral variation. Genetics 155 1415–1427
M Kimura (1983) The neutral theory of molecular evolution. Cambridge University Press Cambridge
DA Kirby W Stephan (1996) ArticleTitleMulti-locus selection and the structure of variation at the white gene of Drosophila melanogaster. Genetics 144 635–645 Occurrence Handle1:CAS:528:DyaK28XmsVanu7o%3D Occurrence Handle8889526
RC Lewontin (1974) The genetic basis of evolutionary change. Columbia University Press New York
JS Liu (2001) Monte Carlo strategies in scientific computing. Springer New York
J Maynard Smith J Haigh (1974) ArticleTitleThe hitch-hiking effect of a favorable gene. Genet Res 23 23–35 Occurrence Handle4407212
JH McDonald M Kreitman (1991) ArticleTitleAdaptive protein evolution at the Adh locus in Drosophila. Nature 351 652–654 Occurrence Handle1:CAS:528:DyaK3MXks1yrtLs%3D Occurrence Handle1904993
N Metropolis A Rosenbluth M Rosenbluth A Teller E Teller (1953) ArticleTitleEquations of state calculations by fast computing machines. J Chem Phys 21 1087–1091 Occurrence Handle1:CAS:528:DyaG3sXltlKhsw%3D%3D
R Nielsen Z Yang (1998) ArticleTitleLikelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148 929–936 Occurrence Handle1:CAS:528:DyaK1cXks1eitr8%3D Occurrence Handle9539414
DI Nurminsky (2001) ArticleTitleGenes in sweeping competition. Cell Mol Life Sci 58 125–134 Occurrence Handle1:CAS:528:DC%2BD3MXhslKnsrs%3D Occurrence Handle11229811
T Ohta (1992) ArticleTitleThe nearly neutral theory of molecular evolution. Annu Rev Ecol Syst 23 263–286 Occurrence Handle10.1146/annurev.es.23.110192.001403
SA Sawyer DL Hartl (1992) ArticleTitlePopulation genetics of polymorphism and divergence. Genetics 132 1161–1176
KJ Schmid D Tautz (1997) ArticleTitleA screen for fast evolving genes from Drosophila. Proc Natl Acad Sci USA 94 9746–9750 Occurrence Handle10.1073/pnas.94.18.9746 Occurrence Handle1:CAS:528:DyaK2sXlvVWit7Y%3D Occurrence Handle9275195
NGC Smith A Eyre-Walker (2002) ArticleTitleAdaptive protein evolution in Drosophila. Nature 415 1022–1024 Occurrence Handle10.1038/4151022a Occurrence Handle1:CAS:528:DC%2BD38XhvVWjsLs%3D Occurrence Handle11875568
W Stephan (1996) ArticleTitleThe rate of compensatory evolution. Genetics 144 419–426 Occurrence Handle1:STN:280:ByiD3s3psFM%3D Occurrence Handle8878705
WJ Swanson AG Clark HM Waldrip-Dail MF Wolfner CF Aquadro (2001) ArticleTitleEvolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci USA 98 7375–7379 Occurrence Handle10.1073/pnas.131568198 Occurrence Handle11404480
F Tajima (1989) ArticleTitleStatistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595 Occurrence Handle1:CAS:528:DyaK3cXhslentA%3D%3D Occurrence Handle2513255
AR Templeton (1998) ArticleTitleNested clade analysis of phylogeographical data: Testing hypotheses about gene flow and population history. Mol Ecol 7 381–397 Occurrence Handle1:CAS:528:DyaK1cXjs1Siu78%3D Occurrence Handle9627999
AR Templeton (2002) ArticleTitleOut of Africa again and again. Nature 416 45–51 Occurrence Handle1:CAS:528:DC%2BD38XitlSitr0%3D Occurrence Handle11882887
J Wakeley (2000) ArticleTitleThe effects of subdivision on the genetic divergence of populations and species. Evol Int J Org Evol 54 1092–1101 Occurrence Handle1:CAS:528:DC%2BD3cXntVylt7Y%3D
Z Yang (1998) ArticleTitleLikelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15 568–573 Occurrence Handle9580986
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sawyer, S.A., Kulathinal, R.J., Bustamante, C.D. et al. Bayesian Analysis Suggests that Most Amino Acid Replacements in Drosophila Are Driven by Positive Selection . J Mol Evol 57 (Suppl 1), S154–S164 (2003). https://doi.org/10.1007/s00239-003-0022-3
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s00239-003-0022-3