Abstract
Purpose
The aim of this study was to evaluate the performance of the NONMEM prior functionality compared to a full Bayesian method when applied to population physiological models using diazepam as a case study.
Methods
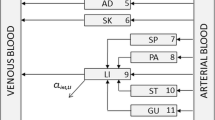
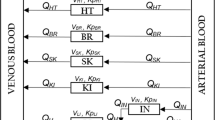
Whole-body physiologically based pharmacokinetic (WBPBPK) models for diazepam were initially developed, tested and calibrated for rats and man using a full Bayesian analysis as implemented in WinBUGS. The final models were implemented in NONMEM and the results from the two analyses compared in terms of parameter estimates, measures of parameter precision and run times.
Results
NONMEM population parameter estimates were in close agreement with those produced by the Bayesian analysis although there was a substantial shortening of run time for both the animal WBPBPK model (4.5 vs. 21 h) and human WBPBPK models (2 vs. 167 h). The adequacy of the model and the final parameter estimates were judged to be sufficient by the model’s ability to describe individual tissue concentration-time profiles. The model provided a good overall description of the plasma concentration-time data in both rat and man with comparable parameter precision. A limited nonparametric bootstrap (n = 50) was performed to assess parameter sensitivity, bias and imprecision. No systematic bias was seen when comparing bootstrap means to final parameter estimates.
Conclusions
The ease of implementation and reductions in run time hopefully provide a further step forward in allowing the wider use of these complex and information-rich models together with clinical data in the future.




Similar content being viewed by others
References
Nestorov I (2003) Whole body pharmacokinetic models. Clin Pharmacokinet 42:883–908
Gueorguieva I, Aarons L, Rowland M (2006) Diazepam pharmacokinetics from preclinical to Phase I using a Bayesian population physiological model with informative prior distributions in WinBUGS. J Pharmacokinet Pharmacodyn 33:571–594
Gisleskog PO, Karlsson MO, Beal SL (2002) Use of prior information to stabilize a population data analysis. J Pharmacokinet Pharmacodyn 29:473–505
Duffull SB, Kirkpatrick CM, Green B, Holford NH (2005) Analysis of population pharmacokinetic data using NONMEM and WinBUGS. J Biopharm Stat 15:53–73
Beerli P (2006) Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22:341–345
Lee P (1997) Bayesian statistics: an introduction, 2nd edn. Wiley, London
Gueorguieva I, Nestorov I, Rowland M (2004) Fuzzy simulation of pharmacokinetic models: case study of whole body physiologically based model of diazepam. J Pharmacokinet Pharmacodyn 31:185–213
Kuwahira I, Gonzalez NC, Heisler N, Piper J (1993) Regional blood flow in conscious resting rats determined by microsphere distribution. J Appl Physiol 74:203–210
Poulin P, Theil FP (2002) Prediction of pharmacokinetics prior to in vivo studies. 1. Mechanism-based prediction of volume of distribution. J Pharm Sci 91:129–156
Gueorguieva I, Nestorov IA, Murby S, Gisbert S, Collins B, Dickens K, Duffy J, Hussain Z, Rowland M (2004) Development of a whole body physiologically based model to characterise the pharmacokinetics of benzodiazepines. 1: Estimation of rat tissue-plasma partition ratios. J Pharmacokinet Pharmacodyn 31:269–298
Greenblatt DJ, Allen MD, Harmatz JS, Shader RI (1980) Diazepam disposition determinants. Clin Pharmacol Ther 27:301–312
Igari Y, Sugiyama Y, Sawada Y, Iga T, Hanano M (1983) Prediction of diazepam disposition in the rat and man by a physiologically based pharmacokinetic model. J Pharmacokinet Biopharm 11:577–593
Beal SL, Sheiner LB (1992) NONMEM users guide – part VII: conditional estimation methods. NONMEM Project Group, San Francisco
Aarons L (2005) Physiologically based pharmacokinetic modelling: a sound mechanistic basis is needed. Br J Clin Pharmacol 60:581–583
Brown RP, Delp MD, Lindstedt SL, Rhomberg LR, Beliles RP (1997) Physiological parameter values for physiologically based pharmacokinetic models. Toxicol Ind Health 13:407–484
Acknowledgements
Grant Langdon was financially supported by Pfizer Central Research, Sandwich, UK. Data used in the analysis were kindly supplied by the Centre for Applied Pharmacokinetic Research, Manchester.
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A: NONMEM control stream with prior implementation





Appendix B: NONMEM prior subroutine

Rights and permissions
About this article
Cite this article
Langdon, G., Gueorguieva, I., Aarons, L. et al. Linking preclinical and clinical whole-body physiologically based pharmacokinetic models with prior distributions in NONMEM . Eur J Clin Pharmacol 63, 485–498 (2007). https://doi.org/10.1007/s00228-007-0264-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00228-007-0264-x