Heading
Abstract
Aims. To determine whether the variability in cytochrome P450 (CYP)3A4 metabolic function is exhibited at both transcription and translation levels and to examine the population distribution of CYP3A4 enzyme kinetics, protein, and mRNA.
Methods. Enzyme kinetics of testosterone 6β-hydroxylation, immunoblot CYP3A4 protein, and CYP3A4 mRNA were determined in a microsomal bank of human livers. The distribution of these determinations was analyzed using cumulative distribution (probit) plots and normality test variable (NTV) to detect deviation from normality.
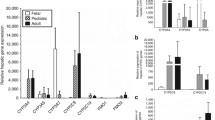
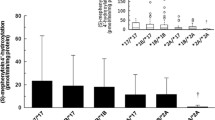
Results. Mean hepatic CYP3A4 protein and relative CYP3A4 mRNA were 35±23 pmol/mg and 79±59 (CYP3A4/β-actin), respectively. Kinetic parameter estimates of testosterone 6β-hydroxylation were 611±684 pmol/mg/min for maximum rate of the reaction (Vmax) and 206±48 µM for the Michaelis constant (Km). The CYP3A4 gene expression and its activity exhibited a relatively high degree of interindividual variability. Furthermore, significant correlation between CYP3A4 protein and Vmax of testosterone 6β-hydroxylation (r=0.82, P<0.001) as well as CYP3A4 protein and its mRNA (r=0.52, P<0.01) was observed. Cumulative distribution plots and histograms for the CYP3A4 protein, its mRNA, and maximum testosterone 6β-hydroxylation exhibited evidences of deviation from log-normal distribution. The minimum NTV value for the distribution of CYP3A4 protein corresponding to the inflection point in the probit occurred at approximately 10% cumulative frequency. The percentage of low CYP3A4 protein phenotype was consistent for CYP3A4 activity and its mRNA. In contrast, the distribution of Km of testosterone 6β-hydroxylation does not show evidence of bimodality.
Conclusions. The distribution of CYP3A4 metabolic function, protein, and mRNA is non-normal and may represent a regulatory polymorphism in hepatic CYP3A4 gene expression. In contrast, assessment of the possibility of a structural variant of CYP3A4 through evaluation of the standard deviation relative to the mean kinetic constant value suggests that structural mutation of CYP3A4 may not be a major factor affecting interindividual variation in CYP3A4 metabolic function.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Accepted in revised form: 15 May 2002
Electronic Publication
Rights and permissions
About this article
Cite this article
Sy, S.K., Ciaccia, A., Li, W. et al. Modeling of human hepatic CYP3A4 enzyme kinetics, protein, and mRNA indicates deviation from log-normal distribution in CYP3A4 gene expression. Eur J Clin Pharmacol 58, 357–365 (2002). https://doi.org/10.1007/s00228-002-0487-9
Received:
Issue Date:
DOI: https://doi.org/10.1007/s00228-002-0487-9