Abstract
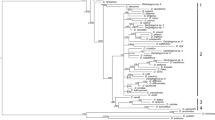
In host–parasite coevolution, parasite innovations including the acquisition of new habitats and novel traits can trigger evolutionary breakthroughs and enhance parasite diversification via accumulation of new hosts. All species of the family Cymothoidae are obligate fish parasites, attaching to exterior body surfaces of fish, the buccal or opercular cavities, or burrowing into abdominal muscle tissue. In the present study, we constructed a molecular phylogeny of 27 cymothoid species that parasitise 38 fish species, combined with 2 prior cymothoid datasets, based on the sequences of mitochondrial 16S rRNA and COI genes. We explored the evolution of the host attachment mode, and the habitat shift from saline water to freshwater. Our evolutionary trees include two freshwater clades, an abdominal burrower clade, and cymothoid clades that are closer to the base of Cymothoidae than those initially analysed. We found that the basal clade of Cymothoidae was Elthusa sacciger, which is parasitic in the opercular cavity of synaphobranchid eels. This result suggests that cymothoids may have originated in deep seas, subsequently expanded to shallow seas, and then to brackish and/or freshwater, by shifting host species. Invasion of freshwater habitats has occurred at least twice; freshwater abdominal muscle burrowers living on armoured catfish constitute a clade allied to E. sacciger. The ancestral host attachment site, based on our dataset, was the opercular cavity, followed (sequentially) by buccal colonisation and attachment to the external body.




Similar content being viewed by others
References
Adlard RD, Lester RJG (1995) The life-cycle and biology of Anilocra pomacentri (Isopoda, Cymothoidae), an ectoparasitic isopod of the coral-reef fish, Chromis nitida (Perciformes, Pomacentridae). Aust J Zool 43:271–281. doi:10.1071/ZO9950271
Boyko CB, Moss J, Williams JD, Shields JD (2013) A molecular phylogeny of Bopyroidea and Cryptoniscoidea (Crustacea: Isopoda). Syst Biodivers 11:495–506. doi:10.1080/14772000.2013.865679
Bruce NL (1983) Aegidae (Isopoda: Crustacea) from Australia with descriptions of three new species. J Nat Hist 17:757–788. doi:10.1080/00222938300770591
Bruce NL (1986) Revision of the isopod crustacean genus Mothocya Costa, in Hope, 1851 (Cymothoidae: Flabellifera), parasitic on marine fishes. J Nat Hist 20:1089–1189
Bruce NL (1987) Australian Pleopodias Richardson, 1910, and Anilocra Leach, 1818 (Isopoda: Cymothoidae), crustacean parasites of marine fishes. Rec Aust Mus 39:85–130
Bruce NL (1990) The genera Catoessa, Elthusa, Enispa, Ichthyoxenus, Idusa, Livoneca and Norileca n.gen. (Isopoda, Cymothoidae), crustacean parasites of marine fishes, with descriptions of eastern Australian species. Rec Aust Mus 42:247–300. doi:10.3853/j.0067-1975.42.1990.118
Bruce NL (2009) The marine fauna of New Zealand: Isopoda, Aegidae (Crustacea). NIWA Biodivers Mem 122:1–252
Bruce NL, Schotte M (2008) Cymothoidae. In: Boyko CB, Bruce NL, Merrin KL, Ota Y, Poore GCB, Taiti S, Schotte M, Wilson GDF (eds) World Marine, Freshwater and Terrestrial Isopod Crustaceans database. http://www.marinespecies.org/aphia.php?p=taxdetails&id=118274. Accessed 13 Dec 2016. (2008 onwards)
Brusca RC (1981) A monograph on the Isopoda Cymothoidae (Crustacea) of the eastern Pacific. Zool J Linn Soc 73:117–199
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi:10.1093/bioinformatics/btp348
Chilton C (1926) Zoological results of a tour in the far East. The Tanaidacea and Isopoda of Tale Sap. Rec Indian Mus 28:173–185
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fryer G (1968) A new parasitic isopod of the family Cymothoidae from clupeid fishes of Lake Tanganyika: a further Lake Tanganyika enigma. J Zool 156:35–43
Goto R, Kawakita A, Ishikawa H, Hamamura Y, Kato M (2012) Molecular phylogeny of the bivalve superfamily Galeommatoidea (Heterodonta, Veneroida) reveals dynamic evolution of symbiotic lifestyle and interphylum host switching. BMC Evol Biol 12:1–13. doi:10.1186/1471-2148-12-172
Hadfield KA (2012) The biodiversity and systematics of marine fish parasitic isopods of the family Cymothoidae from Southern Africa. Dissertation, University of Johanesburg
Hadfield KA, Bruce NL, Smit NJ (2015) Review of Mothocya Costa in Hope 1851 Crustacea Isopoda Cymothoidae from southern Africa, with the description of a new species. Afr Zool 50:147–163. doi:10.1080/15627020.2015.1043943
Hassanin A (2006) Phylogeny of Arthropoda inferred from mitochondrial sequences: strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol Phylogenet Evol 38:100–116. doi:10.1016/j.ympev.2005.09.012
Horton T, Okamura B (2003) Post-haemorrhagic anaemia in sea bass, Dicentrarchus labrax (L.), caused by blood feeding of Ceratothoa oestroides (Isopoda: Cymothoidae). J Fish Dis 26:401–406. doi:10.1046/j.1365-2761.2003.00476.x
Inoue JG, Miya M, Miller MJ, Sado T, Hanel R, Hatooka K, Aoyama J, Minegishi Y, Nishida M, Tsukamoto K (2010) Deep-ocean origin of the freshwater eels. Biol Lett 6:363–366. doi:10.1098/rsbl.2009.0989
Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M (2015) Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci USA 112:14918–14923. doi:10.1073/pnas.1507669112
Johnson GD, Ida H, Sakaue J, Sado T, Asahida T, Miya M (2012) A ‘living fossil’ eel (Anguilliformes: Protoanguillidae, fam. nov.) from an undersea cave in Palau. Proc R Soc B 279:934–943. doi:10.1098/rspb.2011.1289
Jones CM, Miller TL, Grutter AS, Cribb TH (2008) Natatory-stage cymothoid isopods: description, molecular identification and evolution of attachment. Int J Parasitol 38:477–491. doi:10.1016/j.ijpara.2007.07.013
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi:10.1093/molbev/mst010
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi:10.1093/bioinformatics/bts199
Ketmaier V, Joyce DA, Horton T, Mariani S (2008) A molecular phylogenetic framework for the evolution of parasitic strategies in cymothoid isopods (Crustacea). J Zool Syst Evol Res 46:19–23. doi:10.1111/j.1439-0469.2007.00423.x
Lincoln R (1972) A new species of Lironeca (Isopoda; Cymothoidae) parasitic on cichlid fishes in Lake Tanganyika. Bull Br Mus Nat Hist Zool 21:329–337
Maddison WP, Maddison DR (2015) Mesquite: a modular system for evolutionary analysis version 3.04. http://mesquiteproject.org. Accessed 13 Dec 2016
Martin MB (2015) Taxonomy and phylogeny of the buccal-attaching Cymothoidae (Crustacea: Isopoda) of Australia. Dissertation, University of Tasmania
Mindell D, Sorenson M, Dimcheff D, Hasegawa M, Ast J, Yuri T (1999) Interordinal relationships of birds and other reptiles based on whole mitochondrial genomes. Syst Biol 48:138–152
Nair GA, Nair NB (1983) Effect of infestation with the isopod, Alitropus typus M. Edwards (Crustacea: Flabellifera: Aegidae) on the haematological parameters of the host fish, Channa striatus (Bloch). Aquaculture 30:11–19. doi:10.1016/0044-8486(83)90147-3
Nakabo T (2013) Fishes of Japan with pictorial keys to the species. Tokai University Press, Kanagawa
Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL (2012) Resolution of ray-finned fish phylogeny and timing of diversification. Proc Natl Acad Sci USA 109:13698–13703. doi:10.1073/pnas.1206625109
Nelson JS, Grande TC, Wilson MVH (2016) Fishes of the world. Wiley, Hoboken
Phillips MJ, Penny D (2003) The root of the mammalian tree inferred from whole mitochondrial genomes. Mol Phylogenet Evol 28:171–185. doi:10.1016/S1055-7903(03)00057-5
Pillai NK (1967) Littoral and parasitic isopods from Kerala: families Eurydicidae, Corallanidae and Aegidae-2. J Bombay Nat Hist Soc 64:267–283
Ricklefs RE, Outlaw DC, Svensson-Coelho M, Medeiros MCI, Ellis VA, Latta S (2014) Species formation by host shifting in avian malaria parasites. Proc Natl Acad Sci USA 111:14816–14821. doi:10.1073/pnas.1416356111
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029
Rosenthal A, Coutelle O, Craxton M (1993) Large-scale production of DNA sequencing templates by microtitre format PCR. Nucl Acids Res 21:173–174. doi:10.1093/nar/21.1.173
Saunders B (2012) Discovery of Australia’s fishes: a history of Australian ichthyology to 1930. CSIRO Publishing Collingwood, VIC
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Floors P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701
Smit NJ, Bruce NL, Hadfield KA (2014) Global diversity of fish parasitic isopod crustaceans of the family Cymothoidae. Int J Parasitol Parasit Wildl 3:188–197. doi:10.1016/j.ijppaw.2014.03.004
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi:10.1093/bioinformatics/btu033
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Tanabe AS (2011) Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour 11:914–921. doi:10.1111/j.1755-0998.2011.03021.x
Tarrío R, Rodríguez-Trelles F, Ayala FJ (2001) Shared nucleotide composition biases among species and their impact on phylogenetic reconstructions of the Drosophilidae. Mol Biol Evol 18:1464–1473. doi:10.1093/oxfordjournals.molbev.a003932
Thatcher VE (2006) Isopoda. In: Thatcher VE (ed) Amazon fish parasites. Pensoft Pub, Sofia, pp 416–453
Trilles J-P, Hipeau-Jacquotte R (2012) Symbiosis and parasitism in the Crustacea. In: Forest J, von Vaupel Klein JC (eds) Treatise on zoology—anatomy, taxonomy, biology. The Crustacea. Brill, Leiden, pp 239–317. doi:10.1163/9789004188259_006
Tsai M-L, Dai C-F (1999) Ichthyoxenus fushanensis, new species (Isopoda: Cymothoidae), parasite of the fresh-water fish Varicorhinus barbatulus from northern Taiwan. J Crustacean Biol 19:917–923. doi:10.1163/193724099X00600
Wilson GDF, Paterson JR, Kear BP (2011) Fossil isopods associated with a fish skeleton from the Lower Cretaceous of Queensland, Australia—direct evidence of a scavenging lifestyle in Mesozoic Cymothoida. Palaeontology 54:1053–1068. doi:10.1111/j.1475-4983.2011.01095.x
Yamano H, Yamauchi T, Hosoya K (2011) A new host record of Ichthyoxenus amurensis (Crustacea: Isopoda: Cymothoidae) from the Amur bitterling Rhodeus sericeus (Cypriniformes: Cyprinidae). Limnology 12:103–106. doi:10.1007/s10201-010-0325-1
Zietara MS, Lumme J (2002) Speciation by host switch and adaptive radiation in a fish parasite genus Gyrodactylus (monogenea, gyrodactylidae). Evolution 56:2445–2458. doi:10.1111/j.0014-3820.2002.tb00170.x
Acknowledgements
We sincerely thank J. Akahane, G. Akinaga, S. N. Chiba, R. Dohi, A. Fujii, K. Goto, H. Hamaoka, T. Hattori, T. Hayakawa, N. Hayashida, I. Hirabayashi, H. Hirasaka, S. Isozaki, G. Itani, M. Ito, M. Ito, H. Matsuba, Kanagawa Prefectural Museum of Natural History, H. Katahira, E. Katayama, Y. Kimura, K. Koeda, T. Kunishima, H. Mishima, K. Miyamoto, Y. Miyazaki, T. Moritaki, F. Nakao, N. Nakayama, M. Nishiguchi, M. Ogishi, J. Ohtomi, T. Oka, T. Okada, I. Oura, K. Sakai, T. Sakai, T. Sakumoto, M. Sakurai, T. Sato, T. P. Satoh, Seikai National Fisheries Research Institute, H. Senou, T. Shigeta, S. Sodeyama, R. Shimomura, T. Shinonome, K. Tachihara, R. Tagashira, M. Takami, T. Takaya, F. Tashiro, Toba Aquarium, Tohoku National Fisheries Research Institute, T. Tsukayama, A. Umemoto, M. Utsunomiya, Y. Yagi, A. Yasui, and S. Yoshimi for providing specimens and/or assisting sample collection. Photos in Figs. 1, 4 courtesy of I. Seidel and H. Hirasaka. We are also grateful to members of the Maneno Team (Tanganyika Research Project Team) and staffs of Lake Tanganyika Research Unit, Mpulungu, Zambia, for their support. Research in Lake Tanganyika was conducted under permission from the Zambian Ministry of Agriculture, Food, and Fisheries. This study was supported by Showa Seitoku Memorial Foundation (Grant No. H26) and the Mikimoto Fund for marine Ecology (Grant No. H24).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical standard
All experiments were conducted in accordance with the Ethical Guidelines for Animal Experiments of Ehime University.
Additional information
Responsible Editor: T. Reusch.
Reviewed by C. Boyko and an undisclosed expert.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hata, H., Sogabe, A., Tada, S. et al. Molecular phylogeny of obligate fish parasites of the family Cymothoidae (Isopoda, Crustacea): evolution of the attachment mode to host fish and the habitat shift from saline water to freshwater. Mar Biol 164, 105 (2017). https://doi.org/10.1007/s00227-017-3138-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-017-3138-5