Abstract
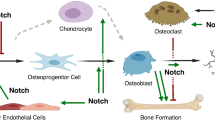
Notch signaling mediates cell-to-cell interactions that are critical for embryonic development and tissue renewal. In the canonical signaling pathway, the Notch receptor is cleaved following ligand binding, resulting in the release and nuclear translocation of the Notch intracellular domain (NICD). NICD induces gene expression by forming a ternary complex with the DNA binding protein CBF1/Rbp-Jk, Suppressor of Hairless, Lag1, and Mastermind-Like (Maml). Hairy Enhancer of Split (Hes) and Hes related with YRPW motif (Hey) are classic Notch targets. Notch canonical signaling plays a central role in skeletal development and bone remodeling by suppressing the differentiation of skeletal cells. The skeletal phenotype of mice misexpressing Hes1 phenocopies partially the effects of Notch misexpression, suggesting that Hey proteins mediate most of the skeletal effects of Notch. Dysregulation of Notch signaling is associated with diseases affecting human skeletal development, such as Alagille syndrome, brachydactyly and spondylocostal dysostosis. Somatic mutations in Notch receptors and ligands are found in tumors of the skeletal system. Overexpression of NOTCH1 is associated with osteosarcoma, and overexpression of NOTCH3 or JAGGED1 in breast cancer cells favors the formation of osteolytic bone metastasis. Activating mutations in NOTCH2 cause Hajdu-Cheney syndrome, which is characterized by skeletal defects and fractures, and JAG1 polymorphisms, are associated with variations in bone mineral density. In conclusion, Notch is a regulator of skeletal development and bone remodeling, and abnormal Notch signaling is associated with developmental and postnatal skeletal disorders.

Similar content being viewed by others
References
Olsen BR, Reginato AM, Wang W (2000) Bone development. Annu Rev Cell Dev Biol 16:191–220
Canalis E, Giustina A, Bilezikian JP (2007) Mechanisms of anabolic therapies for osteoporosis. N Engl J Med 357:905–916
D’Souza B, Miyamoto A, Weinmaster G (2008) The many facets of notch ligands. Oncogene 27:5148–5167
Kopan R, Ilagan MX (2009) The canonical notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233
Fortini ME (2009) Notch signaling: the core pathway and its posttranslational regulation. Dev Cell 16:633–647
Iso T, Kedes L, Hamamori Y (2003) HES and HERP families: multiple effectors of the notch signaling pathway. J Cell Physiol 194:237–255
Heitzler P (2010) Biodiversity and noncanonical notch signaling. Curr Top Dev Biol 92:457–481
Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL (2008) NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem 283:6509–6518
Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, O’Keefe RJ, Hilton MJ (2010) RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development 137:1461–1471
Tao J, Chen S, Yang T, Dawson B, Munivez E, Bertin T, Lee B (2010) Osteosclerosis owing to Notch gain of function is solely Rbpj-dependent. J Bone Miner Res 25:2175–2183
McCright B, Lozier J, Gridley T (2006) Generation of new Notch2 mutant alleles. Genesis 44:29–33
Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T (1994) Notch1 is essential for postimplantation development in mice. Genes Dev 8:707–719
Bellavia D, Checquolo S, Campese AF, Felli MP, Gulino A, Screpanti I (2008) Notch3: from subtle structural differences to functional diversity. Oncogene 27:5092–5098
Monet M, Domenga V, Lemaire B, Souilhol C, Langa F, Babinet C, Gridley T, Tournier-Lasserve E, Cohen-Tannoudji M, Joutel A (2007) The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Hum Mol Genet 16:982–992
Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, Smith GH, Stark KL, Gridley T (2000) Notch signaling is essential for vascular morphogenesis in mice. Genes Dev 14:1343–1352
Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ (2002) Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis 33:77–80
Zanotti S, Canalis E (2010) Notch and the skeleton. Mol Cell Biol 30:886–896
Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F (2008) Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med 14:306–314
Mead TJ, Yutzey KE (2009) Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci USA 106:14420–14425
Zanotti S, Smerdel-Ramoya A, Canalis E (2011) Hairy and enhancer of split (HES)1 is a determinant of bone mass. J Biol Chem 286:2648–2657
Canalis E (2005) The fate of circulating osteoblasts. N Engl J Med 352:2014–2016
Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E (2008) Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology 149:3890–3899
Engin F, Yao Z, Yang T, Zhou G, Bertin T, Jiang MM, Chen Y, Wang L, Zheng H, Sutton RE, Boyce BF, Lee B (2008) Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14:299–305
Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, Rowe D (2002) Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res 17:15–25
Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E (2006) Notch 1 overexpression inhibits osteoblastogenesis by suppressing Wnt/beta-catenin but not bone morphogenetic protein signaling. J Biol Chem 281:6203–6210
Zanotti S, Smerdel-Ramoya A, Canalis E (2011) Reciprocal regulation of notch and nuclear factor of activated T-cells (NFAT)c1 transactivation in osteoblasts. J Biol Chem 286:4576–4588
Sitara D, Aliprantis AO (2010) Transcriptional regulation of bone and joint remodeling by NFAT. Immunol Rev 233:286–300
Zhang Y, Lian JB, Stein JL, van Wijnen AJ, Stein GS (2009) The Notch-responsive transcription factor Hes-1 attenuates osteocalcin promoter activity in osteoblastic cells. J Cell Biochem 108:651–659
Lee JS, Thomas DM, Gutierrez G, Carty SA, Yanagawa S, Hinds PW (2006) HES1 cooperates with pRb to activate RUNX2-dependent transcription. J Bone Miner Res 21:921–933
McLarren KW, Lo R, Grbavec D, Thirunavukkarasu K, Karsenty G, Stifani S (2000) The mammalian basic helix loop helix protein HES-1 binds to and modulates the transactivating function of the runt-related factor Cbfa1. J Biol Chem 275:530–538
Shen Q, Christakos S (2005) The vitamin D receptor, Runx2, and the Notch signaling pathway cooperate in the transcriptional regulation of osteopontin. J Biol Chem 280:40589–40598
Salie R, Kneissel M, Vukevic M, Zamurovic N, Kramer I, Evans G, Gerwin N, Mueller M, Kinzel B, Susa M (2010) Ubiquitous overexpression of Hey1 transcription factor leads to osteopenia and chondrocyte hypertrophy in bone. Bone 46:680–694
Tu X, Lim K, Ganss K, Surendran R, Kopan R, Gessler M, Long F (2009) Notch inhibits early stages of osteoblastogenesis through RBP-Jk and hey proteins. J Bone Miner Res 24(Suppl 1). http://www.asbmr.org/Meetings/AnnualMeeting/AbstractDetail.aspx?aid=3796349f-2835-4777-8547-1d873506308f
Teitelbaum SL (2007) Osteoclasts: What do they do and how do they do it? Am J Pathol 170:427–435
Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S (2003) Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood 101:2227–2234
Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, Bigas A, Jimi E, Okabe K (2008) The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol Cell Biol 28:6402–6412
Sethi N, Dai X, Winter CG, Kang Y (2011) Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 19:192–205
Tian J, Ling L, Shboul M, Lee H, O’Connor B, Merriman B, Nelson SF, Cool S, Ababneh OH, Al-Hadidy A, Masri A, Hamamy H, Reversade B (2010) Loss of CHSY1, a secreted FRINGE enzyme, causes syndromic brachydactyly in humans via increased NOTCH signaling. Am J Hum Genet 87:768–778
Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T (1998) Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev 12:1046–1057
Sidow A, Bulotsky MS, Kerrebrock AW, Bronson RT, Daly MJ, Reeve MP, Hawkins TL, Birren BW, Jaenisch R, Lander ES (1997) Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature 389:722–725
Gridley T (2003) Notch signaling and inherited disease syndromes. Hum Mol Genet 12:R9–R13
Krantz ID, Piccoli DA, Spinner NB (1999) Clinical and molecular genetics of alagille syndrome. Curr Opin Pediatr 11:558–564
McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB (2006) NOTCH2 mutations cause alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 79:169–173
McCright B, Lozier J, Gridley T (2002) A mouse model of alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development 129:1075–1082
Turnpenny PD, Alman B, Cornier AS, Giampietro PF, Offiah A, Tassy O, Pourquie O, Kusumi K, Dunwoodie S (2007) Abnormal vertebral segmentation and the notch signaling pathway in man. Dev Dyn 236:1456–1474
Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS (2002) Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129:1795–1806
Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES (1998) The mouse pudgy mutation disrupts delta homologue Dll3 and initiation of early somite boundaries. Nat Genet 19:274–278
Saga Y, Hata N, Koseki H, Taketo MM (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev 11:1827–1839
Cornier AS, Staehling-Hampton K, Delventhal KM, Saga Y, Caubet JF, Sasaki N, Ellard S, Young E, Ramirez N, Carlo SE, Torres J, Emans JB, Turnpenny PD, Pourquie O (2008) Mutations in the MESP2 gene cause spondylothoracic dysostosis/Jarcho-Levin syndrome. Am J Hum Genet 82:1334–1341
Whittock NV, Sparrow DB, Wouters MA, Sillence D, Ellard S, Dunwoodie SL, Turnpenny PD (2004) Mutated MESP2 causes spondylocostal dysostosis in humans. Am J Hum Genet 74:1249–1254
Stanley P (2007) Regulation of Notch signaling by glycosylation. Curr Opin Struct Biol 17:530–535
Dunwoodie SL (2009) Mutation of the fucose-specific beta 1,3N-acetylglucosaminyltransferase LFNG results in abnormal formation of the spine. Biochim Biophys Acta 1792:100–111
Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, Turnpenny PD, Kusumi K, Sillence D, Dunwoodie SL (2006) Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet 78:28–37
Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R (2001) Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev 15:2642–2647
Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL (2008) Mutation of Hairy-and-Enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum Mol Genet 17:3761–3766
Sparrow DB, Sillence D, Wouters MA, Turnpenny PD, Dunwoodie SL (2010) Two novel missense mutations in Hairy-and-Enhancer-of-Split-7 in a family with spondylocostal dysostosis. Eur J Hum Genet 18:674–679
Isidor B, Lindenbaum P, Pichon O, Bezieau S, Dina C, Jacquemont S, Martin-Coignard D, Thauvin-Robinet C, Le MM, Mandel JL, David A, Faivre L, Cormier-Daire V, Redon R, Le CC (2011) Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat Genet 43:306–308
Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, Kim KH, Pauli RM, Aftimos S, Stewart H, Kim CA, Holder-Espinasse M, Robertson SP, Drake WM, Trembath RC (2011) Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet 43:303–305
Kung AW, Xiao SM, Cherny S, Li GH, Gao Y, Tso G, Lau KS, Luk KD, Liu JM, Cui B, Zhang MJ, Zhang ZL, He JW, Yue H, Xia WB, Luo LM, He SL, Kiel DP, Karasik D, Hsu YH, Cupples LA, Demissie S, Styrkarsdottir U, Halldorsson BV, Sigurdsson G, Thorsteinsdottir U, Stefansson K, Richards JB, Zhai G, Soranzo N, Valdes A, Spector TD, Sham PC (2010) Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet 86:229–239
Radtke F, Fasnacht N, MacDonald HR (2010) Notch signaling in the immune system. Immunity 32:14–27
Leong KG, Karsan A (2006) Recent insights into the role of Notch signaling in tumorigenesis. Blood 107:2223–2233
Engin F, Bertin T, Ma O, Jiang MM, Wang L, Sutton RE, Donehower LA, Lee B (2009) Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet 18:1464–1470
Zhang P, Yang Y, Zweidler-McKay PA, Hughes DP (2008) Critical role of notch signaling in osteosarcoma invasion and metastasis. Clin Cancer Res 14:2962–2969
Zhang Z, Wang H, Ikeda S, Fahey F, Bielenberg D, Smits P, Hauschka PV (2010) Notch3 in human breast cancer cell lines regulates osteoblast–cancer cell interactions and osteolytic bone metastasis. Am J Pathol 177:1459–1469
Ryeom SW (2011) The cautionary tale of side effects of chronic Notch1 inhibition. J Clin Invest 121:508–509
Moellering RE, Cornejo M, Davis TN, Del BC, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE (2009) Direct inhibition of the NOTCH transcription factor complex. Nature 462:182–188
Acknowledgement
This study was supported by grant DK045227 (to E. C.) from the National Institute of Diabetes, Digestive and Kidney Diseases and by a Research Fellowship from the Arthritis Foundation (to S. Z.).
Author information
Authors and Affiliations
Corresponding author
Additional information
The authors have stated that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Zanotti, S., Canalis, E. Notch Regulation of Bone Development and Remodeling and Related Skeletal Disorders. Calcif Tissue Int 90, 69–75 (2012). https://doi.org/10.1007/s00223-011-9541-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00223-011-9541-x