Abstract
With its ability to easily infect each step of food production chain, Salmonella spp. stands as one of the most important foodborne pathogens which impose threats against economy and human health by creating high morbidity and mortality, worldwide. Although sensitive screening and detection methodologies based on common conventional techniques have been developed for Salmonella spp., their long turnaround time and strict legislative demands directs the researchers to develop more sensitive and rapid methods. Digital PCR has many potential advantages over its closest competitor, Real-Time PCR. It gives more accurate, sensitive, and precise results, especially for samples that contain low pathogen concentrations and shows more resistance against PCR inhibitors. In this study, we developed a droplet digital PCR screening methodology with a short pre-enrichment step. For this purpose, 25 g raw minced meat was subjected to Salmonella contamination, and following a very short 3 h enrichment step, bacterial isolation from 5 mL of sample was achieved. For the specific detection of the Salmonella spp., bipA gene region was selected as a better candidate. Subsequently, droplet digital PCR setup was performed and detection sensitivity of the test was determined as 1.39 cfu/mL. Furthermore, parallel experiments were also conducted using Real-Time PCR, for comparative and validative purposes. Conclusively, data gathered in this study have shown that droplet digital PCR with our proposed setup can sensitively detect Salmonella spp. in raw minced meat samples while generating the same-day results.









Similar content being viewed by others
References
Besser JM (2018) Salmonella epidemiology: a whirlwind of change. Food Microbiol 71:55–59. https://doi.org/10.1016/j.fm.2017.08.018
Havelaar AH, Kirk MD, Torgerson PR, Gibb HJ, Hald T, Lake RJ, Praet N, Bellinger DC, Silva NR, Gargouri N, Speybroeck N, Cawthorne A, Mathers C, Stein C, Angulo FJ, Devleesschauwer B (2015) World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 12(12):e1001923. https://doi.org/10.1371/journal.pmed.1001923
Blackburn WC, McClure PJ (eds) (2009) Foodborne pathogens: hazards, risk analysis and control. Elsevier, Amsterdam
Mughini-Gras L, Franz E, van Pelt W (2018) New paradigms for Salmonella source attribution based on microbial subtyping. Food Microbiol 71:60–67. https://doi.org/10.1016/j.fm.2017.03.002
McEntire J, Acheson D, Siemens A, Eilert S, Robach M (2014) The public health value of reducing Salmonella levels in raw meat and poultry. Food Protect Trends 34(6):386–392
de Freitas Neto OC, Penha Filho RAC, Barrow P, Berchieri Junior A (2010) Sources of human non-typhoid salmonellosis: a review. Braz J Poultry Sci 12(1):01–11. https://doi.org/10.1590/S1516-635X2010000100001
Gill AO, Gill CO (2015) Developments in sampling and test methods for pathogens in fresh meat. Advances in Microbial Food Safety. Woodhead Publishing, Cambridge, pp 257–280
Carli KT, Unal CB, Caner V, Eyigor A (2001) Detection of Salmonellae in chicken feces by a combination of tetrathionate broth enrichment, capillary PCR, and capillary gel electrophoresis. J Clin Microbiol 39(5):1871–1876. https://doi.org/10.1128/JCM.39.5.1871-1876.2001
McGuinness S, McCabe E, O’Regan E, Dolan A, Duffy G, Burgess C, Fanning S, Barry T, O’Grady J (2009) Development and validation of a rapid real-time PCR based method for the specific detection of Salmonella on fresh meat. Meat Sci 83(3):555–562. https://doi.org/10.1016/j.meatsci.2009.07.004
Hagren V, von Lode P, Syrjälä A, Korpimäki T, Tuomola M, Kauko O, Nurmi J (2008) An 8-hour system for Salmonella detection with immunomagnetic separation and homogeneous time-resolved fluorescence PCR. Int J Food Microbiol 125(2):158–161. https://doi.org/10.1016/j.ijfoodmicro.2008.03.037
Hara-Kudo Y, Yoshino M, Kojima T, Ikedo M (2005) Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol Lett 253(1):155–161. https://doi.org/10.1016/j.femsle.2005.09.032
Wolffs PF, Glencross K, Thibaudeau R, Griffiths MW (2006) Direct quantitation and detection of salmonellae in biological samples without enrichment, using two-step filtration and real-time PCR. Appl Environ Microbiol 72(6):3896–3900. https://doi.org/10.1128/AEM.02112-05
Law JWF, Ab Mutalib NS, Chan KG, Lee LH (2015) Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol 5:770. https://doi.org/10.3389/fmicb.2014.00770
Garrido A, Chapela MJ, Román B, Fajardo P, Lago J, Vieites JM, Cabado AG (2013) A new multiplex real-time PCR developed method for Salmonella spp. and Listeria monocytogenes detection in food and environmental samples. Food Control 30(1):76–85. https://doi.org/10.1016/j.foodcont.2012.06.029
Mangal M, Bansal S, Sharma SK, Gupta RK (2016) Molecular detection of foodborne pathogens: a rapid and accurate answer to food safety. Crit Rev Food Sci Nutr 56(9):1568–1584. https://doi.org/10.1080/10408398.2013.782483
Chapela MJ, Garrido-Maestu A, Cabado AG (2015) Detection of foodborne pathogens by qPCR: a practical approach for food industry applications. Cogent Food Agric 1(1):1013771. https://doi.org/10.1080/23311932.2015.1013771
Hoshino T, Inagaki F (2012) Molecular quantification of environmental DNA using microfluidics and digital PCR. Syst Appl Microbiol 35(6):390–395. https://doi.org/10.1016/j.syapm.2012.06.006
Nixon G, Garson JA, Grant P, Nastouli E, Foy CA, Huggett JF (2014) Comparative study of sensitivity, linearity, and resistance to inhibition of digital and nondigital polymerase chain reaction and loop mediated isothermal amplification assays for quantification of human cytomegalovirus. Anal Chem 86(9):4387–4394
Rački N, Dreo T, Gutierrez-Aguirre I, Blejec A, Ravnikar M (2014) Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant methods 10(1):42
Rossen L, Nørskov P, Holmstrøm K, Rasmussen OF (1992) Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol 17(1):37–45. https://doi.org/10.1016/0168-1605(92)90017-W
Zhao Y, Xia Q, Yin Y, Wang Z (2016) Comparison of droplet digital PCR and quantitative PCR assays for quantitative detection of Xanthomonas citri. subsp citri. PLoS ONE 11:7. https://doi.org/10.1371/journal.pone.0159004
Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, Kitano TK et al (2011) High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem 83(22):8604–8610. https://doi.org/10.1021/ac202028g
Huggett JF, Cowen S, Foy CA (2015) Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem 61(1):79–88. https://doi.org/10.1373/clinchem.2014.221366
Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR (2012) Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem 84(2):1003–1011. https://doi.org/10.1021/ac202578x
Bhunia AK (2014) One day to one hour: how quickly can foodborne pathogens be detected? Future Microbiol 9(8):935–946. https://doi.org/10.2217/fmb.14.61
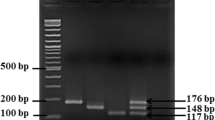
Calvó L, Martínez-Planells A, Pardos-Bosch J, Garcia-Gil LJ (2008) A new real-time PCR assay for the specific detection of Salmonella spp. targeting the bipA gene. Food Anal Methods 1(4):236–242. https://doi.org/10.1007/s12161-007-9008-x
Salehi TZ, Mahzounieh M, Saeedzadeh A (2005) Detection of invA gene in isolated Salmonella from broilers by PCR method. Int J Poult Sci 4(8):557–559. https://doi.org/10.3923/ijps.2005.557.559
Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, Pfaffl MW et al (2013) The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem 59(6):892–902. https://doi.org/10.1373/clinchem.2013.206375
Demeke T, Dobnik D (2018) Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal Bioanal Chem 410(17):4039–4050. https://doi.org/10.1007/s00216-018-1010-1
Wilson IG (1997) Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63(10):3741
Dreo T, Pirc M, Ramšak Ž, Pavšič J, Milavec M, Žel J, Gruden K (2014) Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: a case study of fire blight and potato brown rot. Anal Bioanal Chem 406(26):6513–6528. https://doi.org/10.1007/s00216-014-8084-1
Pecoraro S, Berben G, Burns M, Corbisier P, De Giacomo M, De Loose M, Dagand E, Dobnik D, Eriksson R, Holst-Jensen A, Kagkli DM, Kreysa J, Lievens A, Mäde D, Mazzara M, Paternò A, Peterseil V, Savini C, Sovová T, Sowa S, Spilsberg B (2019) Overview and recommendations for the application of digital PCR. EUR 29673 EN. Publications Office of the European Union, Luxembourg. https://doi.org/10.2760/192883. ISBN 978-92-7600180-5
Malorny B, Löfström C, Wagner M, Krämer N, Hoorfar J (2008) Enumeration of Salmonella bacteria in food and feed samples by real-time PCR for quantitative microbial risk assessment. Appl Environ Microbiol 74(5):1299–1304. https://doi.org/10.1128/AEM.02489-07
European Commission (2005) Commission Regulation (EC) No 2073/2005 of 15 November 2005 on microbiological criteria for foodstuffs. Off J Eur Union L338:1–26
Fachmann MSR, Löfström C, Hoorfar J, Hansen F, Christensen J, Mansdal S, Josefsen MH (2017) Detection of Salmonella enterica in meat in less than 5 hours by a low-cost and noncomplex sample preparation method. Appl Environ Microbiol 83(5):e03151–e3216. https://doi.org/10.1128/AEM.03151-16
Sedlak RH, Jerome KR (2013) Viral diagnostics in the era of digital polymerase chain reaction. Diagn Microbiol Infect Dis 75(1):1–4. https://doi.org/10.1016/j.diagmicrobio.2012.10.009
Shanmugasamy M, Velayutham T, Rajeswar J (2011) Inv A gene specific PCR for detection of Salmonella from broilers. Veterinary World 4(12):562. https://doi.org/10.5455/vetworld.2011.562-564
Cremonesi P, Cortimiglia C, Picozzi C, Minozzi G, Malvisi M, Luini M, Castiglioni B (2016) Development of a droplet digital polymerase chain reaction for rapid and simultaneous identification of common foodborne pathogens in soft cheese. Front Microbiol 7:1725. https://doi.org/10.3389/fmicb.2016.01725
Porcellato D, Narvhus J, Skeie SB (2016) Detection and quantification of Bacillus cereus group in milk by droplet digital PCR. J Microbiol Methods 127:1–6. https://doi.org/10.1016/j.mimet.2016.05.012
Wang M, Yang J, Gai Z, Huo S, Zhu J, Li J, Zhang L et al (2018) Comparison between digital PCR and real-time PCR in detection of Salmonella typhimurium in milk. Int J Food Microbiol 266:251–256. https://doi.org/10.1016/j.ijfoodmicro.2017.12.011
Acknowledgements
We would like to thank Prof. Fikrettin Şahin from the Yeditepe University, Prof. Işın Akyar from the Acıbadem Labmed, and Prof. Gülay Özcengiz from the Middle East Technical Univeristy for providing strains used in this study.
Funding
This study was funded by The Scientific and Technological Research Council of Turkey (Grant no. 1120262).
Author information
Authors and Affiliations
Contributions
All authors have contributed to the study conception and design. Material preparation, data collection, and analysis were performed by YYÖ, ÖİS, SK, EÖ, CBU, and AYK. The first draft of the manuscript was written by YYÖ and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest regarding the publication of this paper.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Öz, Y.Y., Sönmez, Ö.İ., Karaman, S. et al. Rapid and sensitive detection of Salmonella spp. in raw minced meat samples using droplet digital PCR. Eur Food Res Technol 246, 1895–1907 (2020). https://doi.org/10.1007/s00217-020-03531-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03531-x