Abstract
The continuous development of fast and simple new methods to identify animal-derived ingredients is very important for the authentication of meat products. This study intended to develop a multiplex PCR method using new species-specific nuclear DNA (nDNA) sequences for the detection of ingredients derived from sheep/goat, bovine, chicken, duck and pig in meat products. Sequence alignment analysis in 53 species showed high specificity of species-specific nDNA. Species-specific primers were designed on the conservative region of each species-specific nDNA sequence. The specificity and conservation of the sequences and primers were verified by PCR reaction and sequencing with the limit of detection down to 0.5 ng. Then, a species-specific multiplex PCR method was developed and optimized to simultaneously detect sheep/goat (237 bp), bovine (223 bp), chicken (192 bp), duck (168 bp) and pig (154 bp) in one reaction. Various processed meat products containing one or more animal-derived ingredients were detected by the developed multiplex PCR method, and the results were consistent with their labeled meat species. Our study provides a fast and simple detection method for regulating labeling of animal-derived ingredients in meat products.




Similar content being viewed by others
References
Hsieh YH, Ofori JA (2014) Detection of horse meat contamination in raw and heat-processed meat products. J Agric Food Chem 62(52):12536
Hsieh YHP, Chen FC, Sheu SC (1997) AAES research developing simple, inexpensive tests for meat products. Highlights Agric Res 44(2):19–20
Ayaz Y, Ayaz ND, Erol I (2010) Detection of species in meat and meat products using enzyme-linked immunosorbent assay. J Muscle Foods 17(2):214–220
Doosti A, Ghasemi Dehkordi P, Rahimi E (2014) Molecular assay to fraud identification of meat products. J Food Sci Technol 51(1):148–152
Di Pinto A, Bottaro M, Bonerba E, Bozzo G, Ceci E, Marchetti P et al (2015) Occurrence of mislabeling in meat products using DNA-based assay. J Food Sci Technol 52:2479–2484
Izadpanah M, Mohebali N, Elyasi Gorji Z, Farzaneh P, Vakhshiteh F, Shahzadeh Fazeli SA (2018) Simple and fast multiplex PCR method for detection of species origin in meat products. J Food Sci Technol 55(2):698–703
Sheikha AFE, Mokhtar NFK, Amie C, Lamasudin DU, Isa NM, Mustafa S (2017) Authentication technologies using DNA-based approaches for meats and halal meats determination. Food Biotechnol 31(4):281–315
Guan F, Jin Y-T, Zhao J, Xu A-C, Luo Y-Y (2018) A PCR method that can be further developed into PCR-RFLP assay for eight animal species identification. J Anal Methods Chem. https://doi.org/10.1155/2018/5890140
Teixeira LV, Teixeira CS, Oliveira DAA (2015) Species-specific identification of red-meat and meat derivative products with the PCR-RFLP technique. Arquivo Brasileiro De Medicina Veterinária E Zootecnia 67:309–314
Amaral JS, Santos CG, Melo VS, Costa J, Oliveira MBPP, Mafra I (2015) Identification of duck, partridge, pheasant, quail, chicken and turkey meats by species-specific PCR assays to assess the authenticity of traditional game meat Alheira sausages. Food Control 47:190–195
Prusakova OV, Glukhova XA, Afanas'eva GV, Trizna YA, Nazarova LF, Beletsky IP (2018) A simple and sensitive two-tube multiplex PCR assay for simultaneous detection of ten meat species. Meat Sci 137:34–40
Ahamad MNU, Hossain MM, Uddin SMK, Sultana S, Nizar NNA, Bonny SQ et al (2019) Tetraplex real-time PCR with TaqMan probes for discriminatory detection of cat, rabbit, rat and squirrel DNA in food products. Eur Food Res Technol 245:2183–2194
Köppel R, Ganeshan A, Weber S, Pietsch K, Graf C, Hochegger R et al (2019) Duplex digital PCR for the determination of meat proportions of sausages containing meat from chicken, turkey, horse, cow, pig and sheep. Eur Food Res Technol 245:853–862
Kumar A, Kumar RR, Sharma BD, Gokulakrishnan P, Mendiratta SK, Sharma D (2014) Identification of species origin of meat and meat products on the dna basis: a review. CRC Crit Rev Food Technol. https://doi.org/10.1080/10408398.2012.693978
Hassan B, El-Garhy HAS, Moustafa MMA (2014) Detection of pork adulteration in processed meat by species-specific PCR-QIAxcel procedure based on D-loop and cytb genes. Appl Microbiol Biotechnol 98(23):9805–9816
Hou B, Meng X, Zhang L, Guo J, Li S, Jin H (2015) Development of a sensitive and specific multiplex PCR method for the simultaneous detection of chicken, duck and goose DNA in meat products. Meat Sci 101:90–94
Qin P, Qu W, Xu J, Qiao D, Yao L, Xue F, Chen W (2019) A sensitive multiplex PCR protocol for simultaneous detection of chicken, duck, and pork in beef samples. J Food Sci Technol 56(3):1266–1274
Yasemin D, Pelin U, Senyuva HZ (2012) Detection of porcine DNA in gelatine and gelatine-containing processed food products-Halal/Kosher authentication. Meat Sci 90(3):686–689
Ballin NZ, Vogensen FK, Karlsson AH (2009) Species determination Can we detect and quantify meat adulteration? Meat Sci 83(2):165–174
Floren C, Wiedemann I, Brenig B, Schutz E, Beck J (2015) Species identification and quantification in meat and meat products using droplet digital PCR (ddPCR). Food Chem 173:1054–1058
Köppel R, Ruf J, Zimmerli F, Breitenmoser A (2008) Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey. Eur Food Res Technol 227(4):1199–1203
Laube I, Zagon J, Spiegelberg A, Butschke A, Kroh LW, Broll H (2007) Development and design of a ‘ready-to-use’reaction plate for a PCR-based simultaneous detection of animal species used in foods. Int J Food Sci Technol 42(1):9–17
Xiang W, Shang Y, Wang Q, Xu Y, Zhu P, Huang K, Xu W (2017) Identification of a chicken (Gallus gallus) endogenous reference gene (Actb) and its application in meat adulteration. Food Chem 234:472
Wang W, Liu J, Zhang Q, Zhou X, Liu B (2019) Multiplex PCR assay for identification and quantification of bovine and equine in minced meats using novel specific nuclear DNA sequences. Food Control 105:29–37
Reid GA (1991) Molecular cloning: a laboratory manual. In: Sambrook J, Fritsch EF, Maniatis T (eds) Trends in biotechnology. Cold Spring Harbor Laboratory Press, New York
Kirsten H, Teupser D, Weissfuss J, Wolfram G, Emmrich F, Ahnert P (2007) Robustness of single-base extension against mismatches at the site of primer attachment in a clinical assay. J Mol Med (Berl) 85(4):361–369
Amaral JS, Santos CG, Melo VS, Oliveira MBPP, Mafra I (2014) Authentication of a traditional game meat sausage (Alheira) by species-specific PCR assays to detect hare, rabbit, red deer, pork and cow meats. Food Res Int 60:140–145
Ha J, Kim S, Lee J, Lee S, Lee H, Choi Y, Oh H, Yoon Y (2017) Identification of pork adulteration in processed meat products using the developed mitochondrial DNA-based primers. Korean J Food Sci Anim 37(3):464–468
Tartaglia M, Saulle E, Pestalozza S, Morelli L, Antonucci G, Battaglia PA (1998) Detection of bovine mitochondrial DNA in ruminant feeds: a molecular approach to test for the presence of bovine-derived materials. J Food Prot 61(5):513–518
Xu X, Gullberg A, Arnason U (1996) The complete mitochondrial DNA (mtDNA) of the donkey and mtDNA comparisons among four closely related mammalian species-pairs. J Mol Evol 43(5):438–446
Ali ME, Rahman MM, Hamid SBA, Mustafa S, Bhassu S, Hashim U (2013) Canine-specific pcr assay targeting cytochrome b gene for the detection of dog meat adulteration in commercial frankfurters. Food Anal Methods 7(1):234–241
Wu Q, Xiang S, Wang W, Zhao J, Xia J, Zhen Y et al (2017) Species Identification of Fox-, Mink-, Dog-, and Rabbit-Derived Ingredients by Multiplex PCR and Real-Time PCR Assay. Appl Biochem Biotechnol 185:1–12
Ebbehøj KF, Thomsen PD (1991) Species differentiation of heated meat products by DNA hybridization. Meat Sci 30(3):221
Acknowledgement
This work was supported by the National Science and Technology Major Project of China (2018ZX08012-001-010).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Wenjun Wang declares that he has no conflict of interest. Xiaokang Wang declares that he has no conflict of interest. Qingde Zhang declares that he has no conflict of interest. Zuhong Liu declares that he has no conflict of interest. Xiang Zhou declares that he has no conflict of interest. Bang Liu declares that he has no conflict of interest. We have filed patents for primers to detect bovine and pig specific nuclear DNA sequences.
Compliance with ethics requirements
This article does not contain any studies involving human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
217_2020_3494_MOESM1_ESM.pdf
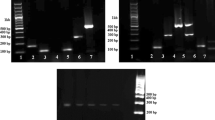
Supplementary file1 Supplementary Fig. 1. All SNPs in the species-specific sequences of target species collected from NCBI, Ensembl and UCSC databases, as well as from the PCR sequencing results of different breeds and individuals. The underlined sequences were primer binding regions; degenerate bases M=A/C, S=G/C, Y=C/T, R=A/G, K=G/T. (PDF 119 kb)
217_2020_3494_MOESM2_ESM.pdf
Supplementary file2 Supplementary Fig. 2. Conservation analysis of PCR for sheep/goat (A), bovine (B), chicken (C), duck (D), and pig (E). In the agarose gel, M, BM2000 DNA Marker, B, blank control, N, negative control, 1-44, samples of different breeds or individuals. (PDF 299 kb)
217_2020_3494_MOESM3_ESM.pdf
Supplementary file3 Supplementary Fig. 3. Detection results in lamb, beef, chicken, duck, pork and mixed meat products. M, BM2000 DNA Marker; B, blank control; N, negative control; P1, positive control of sheep/goat; P2, positive control of bovine; P3, positive control of chicken; P4, positive control of duck; P5, positive control of pig; RLK, roasted lamb kebab; BLR, boiled lamb roll; BH, beef ham; BJ, beef jerky; DCC, deep-fried chicken chop; GCK, grilled chicken kebab; SDN, stewed duck neck; RD, roasted duck; PS, pork sausage; CBS, cooked bacon slices; (a), mixed meat rolls; (b), beef dumplings; (c), beef ham slices; (d), western ham; (e), pork ham slices; (f), sausage; (g), homemade meatball. (PDF 250 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Wang, X., Zhang, Q. et al. A multiplex PCR method for detection of five animal species in processed meat products using novel species-specific nuclear DNA sequences. Eur Food Res Technol 246, 1351–1360 (2020). https://doi.org/10.1007/s00217-020-03494-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-020-03494-z