Abstract
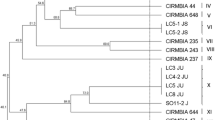
The lactic acid bacteria are very important components involved in the milk products processing. Lactobacillus helveticus, a homofermentative thermophilic lactobacillus commonly occurring in cheeses, has been isolated as the prevailing species in natural koumiss in this work. The species identification of six studied L. helveticus strains was based on rep-PCR fingerprinting with (GTG)5 primer. Biotyping (API 50CH kit, conventional tests) showed phenotypic heterogeneity among isolates and enabled the identification to the genus Lactobacillus only. Ribotyping with restriction enzyme EcoRI yielded a strain-specific restriction patterns and allowed good strain differentiation. Obtained ribotypes of the isolates originating from the koumiss gave unique band patterns with low similarity to the selected reference cultures of L. helveticus. Despite low number of strains analyzed, the ribotype data showed certain heterogeneity that seemed to be not only strain dependent, but also related to the source of isolates. We concluded that analyzed isolates from koumiss represent a new ecovar, L. helveticus ecovar Koumiss.


Similar content being viewed by others
References
Hammes WP, Vogel RF (1995) In: Wood BJB, Holzapfel WH (eds) The lactic acid bacteria, vol 2. The genera of lactic acid bacteria. Blackie Academic and Professional, Glasgow
Callanan M, Kaleta P, O’Callaghan J, O’Sullivan O, Jordan K, McAuliffe O, Sangrador-Vegas A, Slattery L, Fitzgerald GF, Beresford T, Ross RP (2008) Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J Bacteriol 190:727–735
De Los Reyes-Gavilán CG, Limsowtin GKY, Tailliez P, Séchaud L, Accolas J-P (1992) A Lactobacillus helveticus-specific DNA probe detects restriction fragment length polymorphisms in this species. Appl Environ Microbiol 58:3429–3432
Gatti M, Trivisano C, Fabrizi E, Neviani E, Gardini F (2004) Biodiversity among Lactobacillus helveticus strains isolated from different natural whey starter cultures as revealed by classification trees. Appl Environ Microbiol 70:182–190
Mainville I, Robert M, Lee B, Farnworth ER (2006) Polyphasic characterization of the lactic acid bacteria in kefir. Syst Appl Microbiol 29:59–68
Wouters JTM, Ayad EHE, Hugenholtz J, Smit G (2002) Microbes from raw milk for fermented dairy products. Int Dairy J 12:91–109
Narva M, Halleen J, Väänänen K, Korpela R (2004) Effects of Lactobacillus helveticus fermented milk bone cells in vitro. Life Sci 75:1727–1734
Candioti MC, Hynes E, Quiberoni A, Palma SB, Sabbag N, Zalazar CA (2002) Reggianito Argentino Cheese: influence of Lactobacillus helveticus strains isolated from natural whey cultures on cheese making and ripening processes. Int Dairy J 12:923–931
Avila M, Garde S, Medina M, Nunez M (2005) Effect of milk inoculation with bacteriocin-producing lactic acid bacteria on a Lactobacillus helveticus adjunct cheese culture. J Food Prot 68:1026–1033
Hebert EM, De Giori GS, Raya R (2001) Isolation and characterization of a slowly milk-coagulating variant of Lactobacillus helveticus deficient in purine biosynthesis. Appl Environ Microbiol 67:1846–1850
Giraffa G, Gatti M, Rossetti L, Senini L, Neviani E (2000) Molecular diversity within Lactobacillus helveticus as revealed by genotypic characterization. Appl Environ Microbiol 66:1259–1265
Kücükcetin A, Yaygin H, Hinrichs J, Kulozik U (2003) Adaptation of bovine milk towards mares’ milk composition by means of membrane technology for koumiss manufacture. Int Dairy J 13:945–951
Malacarne M, Martuzzi F, Summer A, Mariani P (2002) Protein and fat composition of mare’s milk: some nutritional remarks with reference to human and cow’s milk. Int Dairy J 12:869–877
Viljoen BC (2001) The interaction between yeasts and bacteria in dairy environments. Int J Food Microbiol 69:37–44
Švec P, Sedláček I, Žáčková L, Nováková D, Kukletová M (2009) Lactobacillus spp. associated with early childhood caries. Folia Microbiol 54:53–58
Švec P, Sedláček I, Pantůček R, Devriese L, Doškař J (2001) Evaluation of ribotyping for characterization and identification of Enterococcus haemoperoxidus and Enterococcus moraviensis strains. FEMS Microbiol Lett 203:23–27
Švec P, Dráb V, Sedláček I (2005) Ribotyping of Lactobacillus casei group strains isolated from dairy products. Folia Microbiol 50:223–228
Gevers D, Huys G, Swings J (2001) Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol Lett 205:31–36
Švec P, Ševčíková A, Sedláček I, Bednářová J, Snauwaert C, Lefebvre K, Vandamme P, Vancanneyt M (2007) Identification of lactic acid bacteria isolated from human blood cultures. FEMS Immunol Med Microbiol 49:192–196
Kandler O, Weiss N (1986) In: Sneath PHA, Mair NS, Sharpe ME, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 2. Williams and Wilkins, Baltimore
Fujisawa T, Benno Y, Yaeshima T, Mitsuoka T (1992) Taxonomic study of the Lactobacillus acidophilus group, with recognition of Lactobacillus gallinarum sp. nov. and Lactobacillus johnsonii sp. nov. and synonymy of Lactobacillus acidophilus group A3 (Johnson et al. 1980) with the type strain of Lactobacillus amylovorus (Nakamura 1981). Int J Syst Bacteriol 42:487–491
Naser SM, Hagen KE, Vancanneyt M, Cleenwerck I, Swings J, Tompkins TA (2006) Lactobacillus suntoryeus Cachat and Priest 2005 is a later synonym of Lactobacillus helveticus (Orla-Jensen 1919) Bergey et al. 1925 (Approved Lists 1980). Int J Syst Evol Microbiol 56:355–360
Giraffa G, De Vecchi P, Rossi P, Nicastro G, Fortina MG (1998) Genotypic heterogeneity among Lactobacillus helveticus strains isolated from natural cheese starters. J Appl Microbiol 85:411–416
Quiberoni A, Tailliez P, Quénée P, Suárez V, Reinheimer J (1998) Genetic (RAPD-PCR) and technological diversities among wild Lactobacillus helveticus strains. J Appl Microbiol 85:591–596
Dimitrov Z, Michaylova M, Mincova S (2005) Characterization of Lactobacillus helveticus strains isolated from Bulgarian yoghurt, cheese, plants and human faecal samples by sodium dodecylsulfate polyacrylamide gel electrophoresis of cell-wall proteins, ribotyping and pulsed field gel fingerprinting. Int Dairy J 15:998–1005
Gatti M, Contarini G, Neviani E (1999) Effectiveness of chemometric techniques in discrimination of Lactobacillus helveticus biotypes from natural dairy starter cultures on the basis of phenotypic characteristics. Appl Environ Microbiol 65:1450–1454
Gatti M, Lazzi C, Rossetti L, Mucchetti G, Neviani E (2003) Biodiversity of Lactobacillus helveticus strains present in natural whey starter used for Parmigiano Reggiano cheese. J Appl Microbiol 95:463–470
Watanabe K, Fujimoto J, Sasamoto M, Dugersuren J, Tumursuh T, Shirchin D (2005) Molecular identification of lactic acid bacteria and yeasts isolated from the traditional fermented milk in Mongolia. In: Abstracts, p. 170: third Asian conference for lactic acid bacteria. Bali, Indonesia, 25–26 August 2005
Johnson ME, Steele JL (2001) In: Doyle MP, Beuchat LR, Montville TJ (eds) Food microbiology: fundamentals and frontiers, 2nd edn. ASM Press, Washington DC
Leroy F, De Vuyst L (2004) Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Tech 15:67–78
Acknowledgments
We are grateful to Altanzaya Yansanjav for koumiss samples. This work was supported by the long-term research program MSM0021622416 and project no. 2B08068 of the Ministry of Education, Youth and Sports of the Czech Republic.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sedláček, I., Nováková, D. & Švec, P. Ribotyping and biotyping of Lactobacillus helveticus from the koumiss. Eur Food Res Technol 230, 753–758 (2010). https://doi.org/10.1007/s00217-010-1215-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00217-010-1215-9