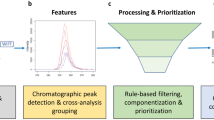
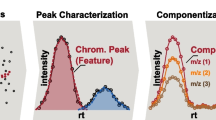
Abstract
Non-targeted analysis (NTA) using high-resolution mass spectrometry allows scientists to detect and identify a broad range of compounds in diverse matrices for monitoring exposure and toxicological evaluation without a priori chemical knowledge. NTA methods present an opportunity to describe the constituents of a sample across a multidimensional swath of chemical properties, referred to as “chemical space.” Understanding and communicating which region of chemical space is extractable and detectable by an NTA workflow, however, remains challenging and non-standardized. For example, many sample processing and data analysis steps influence the types of chemicals that can be detected and identified. Accordingly, it is challenging to assess whether analyte non-detection in an NTA study indicates true absence in a sample (above a detection limit) or is a false negative driven by workflow limitations. Here, we describe the need for accessible approaches that enable chemical space mapping in NTA studies, propose a tool to address this need, and highlight the different ways in which it could be implemented in NTA workflows. We identify a suite of existing predictive and analytical tools that can be used in combination to generate scores that describe the likelihood a compound will be detected and identified by a given NTA workflow based on the predicted chemical space of that workflow. Higher scores correspond to a higher likelihood of compound detection and identification in a given workflow (based on sample extraction, data acquisition, and data analysis parameters). Lower scores indicate a lower probability of detection, even if the compound is truly present in the samples of interest. Understanding the constraints of NTA workflows can be useful for stakeholders when results from NTA studies are used in real-world applications and for NTA researchers working to improve their workflow performance. The hypothetical ChemSpaceTool suggested herein could be used in both a prospective and retrospective sense. Prospectively, the tool can be used to further curate screening libraries and set identification thresholds. Retrospectively, false detections can be filtered by the plausibility of the compound identification by the selected NTA method, increasing the confidence of unknown identifications. Lastly, this work highlights the chemometric needs to make such a tool robust and usable across a wide range of NTA disciplines and invites others who are working on various models to participate in the development of the ChemSpaceTool. Ultimately, the development of a chemical space mapping tool strives to enable further standardization of NTA by improving method transparency and communication around false detection rates, thus allowing for more direct method comparisons between studies and improved reproducibility. This, in turn, is expected to promote further widespread applications of NTA beyond research-oriented settings.





Similar content being viewed by others
References
Place BJ, Ulrich EM, Challis JK, Chao A, Du B, Favela K, et al. An introduction to the benchmarking and publications for non-targeted analysis working group. Anal Chem. 2021;93(49):16289–96.
Peter KT, Phillips AL, Knolhoff AM, Gardinali PR, Manzano CA, Miller KE, et al. Nontargeted analysis study reporting tool: a framework to improve research transparency and reproducibility. Anal Chem [Internet]. 2021;93:13870–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/34618419.
Reymond JL, Ruddigkeit L, Blum L, van Deursen R. The enumeration of chemical space. Wiley Interdiscip Rev Comput Mol Sci. 2012;2(5):717–33.
Knolhoff AM, Premo JH, Fisher CM. A proposed quality control standard mixture and its uses for evaluating nontargeted and suspect screening LC/HR-MS method performance. Anal Chem. 2021;93(3):1596–603.
Pechnick RN. Comparison of the effects of dextromethorphan, dextrorphan, and levorphanol on the hypothalamo-pituitary-adrenal axis. J Pharmacol Exp Ther. 2004;309(2):515–22.
Black G, He G, Denison M, Young T. Using estrogenic activity and nontargeted chemical analysis to identify contaminants in sewage sludge. Environ Sci Technol. 55(10):6729–39.
Léon A, Cariou R, Hutinet S, Hurel J, Guitton Y, Tixier C, et al. HaloSeeker 1.0: a user-friendly software to highlight halogenated chemicals in nontargeted high-resolution mass spectrometry data sets. Anal Chem. 2019;91(5):3500–7.
Koelmel JP, Stelben P, McDonough CA, Dukes DA, Aristizabal-Henao JJ, Nason SL, et al. FluoroMatch 2.0—making automated and comprehensive non-targeted PFAS annotation a reality. Anal Bioanal Chem. 2022;414(3):1201–15.
Loos M, Singer H. Nontargeted homologue series extraction from hyphenated high resolution mass spectrometry data. J Cheminform. 2017;9(1):1–11.
Lowe CN, Isaacs KK, McEachran A, Grulke CM, Sobus JR, Ulrich EM, et al. Predicting compound amenability with liquid chromatography-mass spectrometry to improve non-targeted analysis. Anal Bioanal Chem [Internet]. 2021;413(30):7495–508. Available from: https://doi.org/10.1007/s00216-021-03713-w.
Liigand J, Wang T, Kellogg J, Smedsgaard J, Cech N, Kruve A. Quantification for non-targeted LC/MS screening without standard substances. Sci Rep. 2020;10(1):1–10.
Nuñez JR, McGrady M, Yesiltepe Y, Renslow RS, Metz TO. Chespa: streamlining expansive chemical space evaluation of molecular sets. J Chem Inf Model. 2020;60(12):6251–7.
Acknowledgements
This work was partially supported by the PNNL Laboratory Directed Research and Development program, the m/q Initiative. PNNL is operated by Battelle for the DOE under contract DE-AC05-76RL01830. The participation of Yong-Lai Feng is on behalf of the Government of Canada. The findings and conclusions in this paper have not been formally disseminated by the Food and Drug Administration and should not be construed to represent any agency determination or policy. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the US Environmental Protection Agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest in the publication of this manuscript.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Black, G., Lowe, C., Anumol, T. et al. Exploring chemical space in non-targeted analysis: a proposed ChemSpace tool. Anal Bioanal Chem 415, 35–44 (2023). https://doi.org/10.1007/s00216-022-04434-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-04434-4