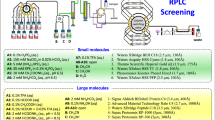
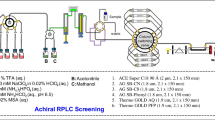
Abstract
Bioprocess development of increasingly challenging therapeutics and vaccines requires a commensurate level of analytical innovation to deliver critical assays across functional areas. Chromatography hyphenated to numerous choices of detection has undeniably been the preferred analytical tool in the pharmaceutical industry for decades to analyze and isolate targets (e.g., APIs, intermediates, and byproducts) from multicomponent mixtures. Among many techniques, ion exchange chromatography (IEX) is widely used for the analysis and purification of biopharmaceuticals due to its unique selectivity that delivers distinctive chromatographic profiles compared to other separation modes (e.g., RPLC, HILIC, and SFC) without denaturing protein targets upon isolation process. However, IEX method development is still considered one of the most challenging and laborious approaches due to the many variables involved such as elution mechanism (via salt, pH, or salt-mediated-pH gradients), stationary phase’s properties (positively or negatively charged; strong or weak ion exchanger), buffer type and ionic strength as well as pH choices. Herein, we introduce a new framework consisting of a multicolumn IEX screening in conjunction with computer-assisted simulation for efficient method development and purification of biopharmaceuticals. The screening component integrates a total of 12 different columns and 24 mobile phases that are sequentially operated in a straightforward automated fashion for both cation and anion exchange modes (CEX and AEX, respectively). Optimal and robust operating conditions are achieved via computer-assisted simulation using readily available software (ACD Laboratories/LC Simulator), showcasing differences between experimental and simulated retention times of less than 0.5%. In addition, automated fraction collection is also incorporated into this framework, illustrating the practicality and ease of use in the context of separation, analysis, and purification of nucleotides, peptides, and proteins. Finally, we provide examples of the use of this IEX screening as a framework to identify efficient first dimension (1D) conditions that are combined with MS-friendly RPLC conditions in the second dimension (2D) for two-dimensional liquid chromatography experiments enabling purity analysis and identification of pharmaceutical targets.
Graphical abstract








Similar content being viewed by others
References
Regalado EL, Haidar Ahmad IA, Bennett R, D’Atri V, Makarov AA, Humphrey GR, et al. The emergence of universal chromatographic methods in the research and development of new drug substances. Acc Chem Res. 2019;52(7):1990–2002.
Johnson DE. Biotherapeutics: challenges and opportunities for predictive toxicology of monoclonal antibodies. Int J Mol Sci. 2018;19(11):3685.
Gautam A, Pan X. The changing model of big pharma: impact of key trends. Drug Discovery Today. 2016;21(3):379–84.
Fekete S, Guillarme D, Sandra P, Sandra K. Chromatographic, electrophoretic, and mass spectrometric methods for the analytical characterization of protein biopharmaceuticals. Anal Chem. 2016;88(1):480–507.
Haidar Ahmad IA, Bennett R, Makey D, Shchurik V, Lhotka H, Mann BF, et al. In silico method development for the reversed-phase liquid chromatography separation of proteins using chaotropic mobile phase modifiers. J Chromatogr B. 2021;1173:122587.
Camperi J, Goyon A, Guillarme D, Zhang K, Stella C. Multi-dimensional LC-MS: the next generation characterization of antibody-based therapeutics by unified online bottom-up, middle-up and intact approaches. Analyst. 2021;146(3):747–69.
Goyon A, D’Atri V, Bobaly B, Wagner-Rousset E, Beck A, Fekete S, et al. Protocols for the analytical characterization of therapeutic monoclonal antibodies. I – non-denaturing chromatographic techniques. J Chromatogr B. 2017;1058:73–84.
Losacco GL, DaSilva JO, Liu J, Regalado EL, Veuthey J-L, Guillarme D. Expanding the range of sub/supercritical fluid chromatography: advantageous use of methanesulfonic acid in water-rich modifiers for peptide analysis. J Chromatogr A. 2021;1642:462048.
Goyon A, Zhang K. Characterization of antisense oligonucleotide impurities by ion-pairing reversed-phase and anion exchange chromatography coupled to hydrophilic interaction liquid chromatography/mass spectrometry using a versatile two-dimensional liquid chromatography setup. Anal Chem. 2020;92(8):5944–51.
Bennett R, Biba M, Liu J, Haidar Ahmad IA, Hicks MB, Regalado EL. Enhanced fluidity liquid chromatography: a guide to scaling up from analytical to preparative separations. J Chromatogr A. 2019;1595:190–8.
Ikegami T. Hydrophilic interaction chromatography for the analysis of biopharmaceutical drugs and therapeutic peptides: a review based on the separation characteristics of the hydrophilic interaction chromatography phases. J Sep Sci. 2019;42(1):130–213.
Jaag S, Shirokikh M, Lämmerhofer M. Charge variant analysis of protein-based biopharmaceuticals using two-dimensional liquid chromatography hyphenated to mass spectrometry. J Chromatogr A. 2021;1636:461786.
Periat A, Fekete S, Cusumano A, Veuthey J-L, Beck A, Lauber M, et al. Potential of hydrophilic interaction chromatography for the analytical characterization of protein biopharmaceuticals. J Chromatogr A. 2016;1448:81–92.
Piestansky J, Barath P, Majerova P, Galba J, Mikus P, Kovacech B, et al. A simple and rapid LC-MS/MS and CE-MS/MS analytical strategy for the determination of therapeutic peptides in modern immunotherapeutics and biopharmaceutics. J Pharm Biomed Anal. 2020;189:113449.
Fekete S, Beck A, Veuthey J-L, Guillarme D. Ion-exchange chromatography for the characterization of biopharmaceuticals. J Pharm Biomed Anal. 2015;113:43–55.
Abou El Azm N, Fleita D, Rifaat D, Mpingirika EZ, Amleh A, El-Sayed MMH. Production of bioactive compounds from the sulfated polysaccharides extracts of Ulva lactuca: post-extraction enzymatic hydrolysis followed by ion-exchange chromatographic fractionation. Molecules. 2019;24(11):2132.
Leblanc Y, Bihoreau N, Chevreux G. Characterization of human serum albumin isoforms by ion exchange chromatography coupled on-line to native mass spectrometry. J Chromatogr B. 2018;1095:87–93.
McGinnis AC, Cummings BS, Bartlett MG. Ion exchange liquid chromatography method for the direct determination of small ribonucleic acids. Anal Chim Acta. 2013;799:57–67.
Mommen GPM, Meiring HD, Heck AJR, de Jong APJM. Mixed-bed ion exchange chromatography employing a salt-free pH gradient for improved sensitivity and compatibility in MudPIT. Anal Chem. 2013;85(14):6608–16.
Bertoletti L, Regazzoni L, Aldini G, Colombo R, Abballe F, Caccialanza G, et al. Separation and characterisation of beta2-microglobulin folding conformers by ion-exchange liquid chromatography and ion-exchange liquid chromatography–mass spectrometry. Anal Chim Acta. 2013;771:108–14.
Haidar Ahmad IA, Shchurik V, Nowak T, Mann BF, Regalado EL. Introducing multifactorial peak crossover in analytical and preparative chromatography via computer-assisted modeling. Anal Chem. 2020;92(19):13443–51.
Ahmed S, Atia NN, Rageh AH. Selectivity enhanced cation exchange chromatography for simultaneous determination of peptide variants. Talanta. 2019;199:347–54.
Verscheure L, Cerdobbel A, Sandra P, Lynen F, Sandra K. Monoclonal antibody charge variant characterization by fully automated four-dimensional liquid chromatography-mass spectrometry. J Chromatogr A. 2021;1653:462409.
Gstöttner C, Klemm D, Haberger M, Bathke A, Wegele H, Bell C, et al. Fast and automated characterization of antibody variants with 4D HPLC/MS. Anal Chem. 2018;90(3):2119–25.
Füssl F, Trappe A, Carillo S, Jakes C, Bones J. Comparative elucidation of cetuximab heterogeneity on the intact protein level by cation exchange chromatography and capillary electrophoresis coupled to mass spectrometry. Anal Chem. 2020;92(7):5431–8.
Schiavone NM, Bennett R, Hicks MB, Pirrone GF, Regalado EL, Mangion I, et al. Evaluation of global conformational changes in peptides and proteins following purification by supercritical fluid chromatography. J Chromatogr B. 2019;1110–1111:94–100.
Li Z, Wang Q, Wang Y, Wang K, Liu Z, Zhang W, et al. An efficient approach based on basic strong cation exchange chromatography for enriching methylated peptides with high specificity for methylproteomics analysis. Anal Chim Acta. 2021;1161:338467.
Patel BA, Pinto NDS, Gospodarek A, Kilgore B, Goswami K, Napoli WN, et al. On-line ion exchange liquid chromatography as a process analytical technology for monoclonal antibody characterization in continuous bioprocessing. Anal Chem. 2017;89(21):11357–65.
Tsay F-R, Haidar Ahmad IA, Henderson D, Schiavone N, Liu Z, Makarov AA, et al. Generic anion-exchange chromatography method for analytical and preparative separation of nucleotides in the development and manufacture of drug substances. J Chromatogr A. 2019;1587:129–35.
Yan Y, Liu AP, Wang S, Daly TJ, Li N. Ultrasensitive characterization of charge heterogeneity of therapeutic monoclonal antibodies using strong cation exchange chromatography coupled to native mass spectrometry. Anal Chem. 2018;90(21):13013–20.
Losacco GL, Wang H, Ahmad IH, DaSilva J, Makarov AA, Mangion I, et al. Enantioselective UHPLC screening combined with in silico modeling for streamlined development of ultrafast enantiopurity assays. Anal Chem. 2022;94(3):1804–12.
Haidar Ahmad IA, Makey DM, Wang H, Shchurik V, Singh AN, Stoll DR, et al. In silico multifactorial modeling for streamlined development and optimization of two-dimensional liquid chromatography. Anal Chem. 2021;93(33):11532–9.
Wang H, Herderschee HR, Bennett R, Potapenko M, Pickens CJ, Mann BF, et al. Introducing online multicolumn two-dimensional liquid chromatography screening for facile selection of stationary and mobile phase conditions in both dimensions. J Chromatogr A. 2020;1622:460895.
Barhate CL, Joyce LA, Makarov AA, Zawatzky K, Bernardoni F, Schafer WA, et al. Ultrafast chiral separations for high throughput enantiopurity analysis. Chem Commun. 2017;53(3):509–12.
Haidar Ahmad IA, Chen W, Halsey HM, Klapars A, Limanto J, Pirrone GF, et al. Multi-column ultra-high performance liquid chromatography screening with chaotropic agents and computer-assisted separation modeling enables process development of new drug substances. Analyst. 2019;144(9):2872–80.
D’Atri V, Murisier A, Fekete S, Veuthey J-L, Guillarme D. Current and future trends in reversed-phase liquid chromatography-mass spectrometry of therapeutic proteins. TrAC Trends Anal Chem. 2020;130:115962.
De Pra M, Greco G, Krajewski MP, Martin MM, George E, Bartsch N, et al. Effects of titanium contamination caused by iron-free high-performance liquid chromatography systems on peak shape and retention of drugs with chelating properties. J Chromatogr A. 2020;1611:460619.
Bennett R, Cohen RD, Wang H, Pereira T, Haverick MA, Loughney JW, et al. A selective plate-based assay for trace EDTA analysis via boron trifluoride-methanol derivatization UHPLC-QqQ-MS/MS enabling biologic and vaccine processes. Anal Chem. 2022;94(3):1678–85.
Guichard N, Fekete S, Guillarme D, Bonnabry P, Fleury-Souverain S. Computer-assisted UHPLC-MS method development and optimization for the determination of 24 antineoplastic drugs used in hospital pharmacy. J Pharm Biomed Anal. 2019;164:395–401.
Haidar Ahmad IA, Kiffer A, Barrientos RC, Losacco GL, Singh A, Shchurik V, Wang H, Mangion I, Regalado EL, et al. In Silico Method Development of Achiral and Chiral Tandem Column Reversed-phase Liquid Chromatography for Multicomponent Pharmaceutical Mixtures. Anal Chem. 2022;94:4065–71.
Makey DM, Shchurik V, Wang H, Lhotka HR, Stoll DR, Vazhentsev A, et al. Mapping the separation landscape in two-dimensional liquid chromatography: blueprints for efficient analysis and purification of pharmaceuticals enabled by computer-assisted modeling. Anal Chem. 2021;93(2):964–72.
Bennett R, Haidar Ahmad IA, DaSilva J, Figus M, Hullen K, Tsay F-R, et al. Mapping the separation landscape of pharmaceuticals: rapid and efficient scale-up of preparative purifications enabled by computer-assisted chromatographic method development. Org Process Res Dev. 2019;23(12):2678–84.
Acknowledgements
The authors would like to thank MRL Postdoctoral Research Fellow Program for sponsoring this research (G.L.L.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Losacco, G.L., Hicks, M.B., DaSilva, J.O. et al. Automated ion exchange chromatography screening combined with in silico multifactorial simulation for efficient method development and purification of biopharmaceutical targets. Anal Bioanal Chem 414, 3581–3591 (2022). https://doi.org/10.1007/s00216-022-03982-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-022-03982-z